【直播】我的基因组(八):原始测序数据质量报告
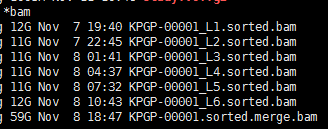
由于我是分期付款,所以我先拿到了我的测序数据的质控结果和比对情况分析报告,需要补齐全款后才能拿到原始测序数据!(中间还出了个小意外,打款的时候不小心多打了30块钱!(⊙o⊙)…不过多打的30块钱想拿回来估计不太可能了,需要填写书面申请表格并且自费快递到公司,这边跨境快递费都不止这个数了) Continue reading
【直播】我的基因组(七):从整体理解全基因组测序数据的变异位点
简单说一下什么是找变异,变异跟突变有什么区别呢?举个栗子:有国际组织规定了人类的参考基因组(如UCSC,ENSEMBL,NCBI等,前面帖子都有讲),就是 AAAAA(这里简化一下,就5个碱基,其实人类基因组多达30亿个) 。现在通过给自己测序得知,我与之对应的是AGCAA,那么我相比国际基因组来说,就是2个变异位点,位于基因组的坐标2和3,但是它们还不能说就是突变。 Continue reading
【直播】我的基因组(六):变异位点注释数据库的准备
通常一个人的全基因组测序数据可以挖掘到四百万个SNVs(跟参考基因组不一样的单碱基位点),还有五十万的indels(insertions or deletions),但是得到的数据通常是以vcf文件格式给出的(自行搜索什么是vcf格式),比如下面:
htseq-counts跟bedtools的区别
我以前写过bedtools和htseq-counts的教程,它们都可以用来对比对好的bam文件进行计数,正好群里有小伙伴问我它们的区别,我就简单做了一个比较,大家可以先看看我以前写的软件教程。写的有的挫:
言归正传,我这里没精力去探究它们的具体原理,只是看看它们数一个read是否属于某个基因的时候,区别在哪里,大家看下图: Continue reading
TPM值就是RPKM的百分比嘛!
很久以前就有人问过这个问题啦,虽然目前主流还是用RPKM/FPKM来形容一个基因的表达量。但是既然大家都说TPM更好,我也来探究一下吧!
我不喜欢看公式,直接说事情,我有一个基因A,它在这个样本的转录组数据中被测序而且mapping到基因组了 5000个的reads,而这个基因A长度是10K,我们总测序文库是50M,所以这个基因A的RPKM值是 5000除以10,再除以50,为10. 就是把基因的reads数量根据基因长度和样本测序文库来normalization 。 Continue reading
仅仅对感兴趣的基因call variation
有这个需求,是因为我们经常对某些细胞系进行有针对性的设计变异,比如BAF155的R1064K呀,H3F3A的K27呀,那我我们拿到高通量测序数据的时候,就肯定希望可以快速的看看这个基因是否被突变成功了。现在比对几乎不耗费什么时间了,但是得到的sam要sort的时候还是蛮耗费时间的。假设,我们已经得到了所有样本的sort好的bam文件,想看看自己设计的基因突变是否成功了,可以有针对性的只call 某个基因的突变!
仔细探究picard的MarkDuplicates 是如何行使去除PCR重复reads功能的
本帖紧跟前面的仔细探究samtools的rmdup是如何行使去除PCR重复reads功能的
同样的我们也是分单端和双端测序来看结果,并且比较两个工具的区别!
首先对于那个单端数据,samtools给出的结果是:[bam_rmdupse_core] 25 / 53 = 0.4717 in library Continue reading
仔细探究samtools的rmdup是如何行使去除PCR重复reads功能的
数据库批量注释不可盲目-annovar数据库错误
我对H3F3A这个基因做了两个突变的cellline,分别是G34V和K27M,现在知道这个基因在hg38上面的坐标是:
Genomic Location for H3F3A Gene
Chromosome: 1
Start:226,061,851 bp from pter End:226,072,002 bp from pter
Size:10,152 bases Orientation:Plus strand
然后我用samtools结合bcftools把该基因区域的snp位点call出来:
samtools mpileup -r chr1:226061851-226072001 -t "DP4" -ugf ~/reference/genome/hg38/hg38.fa *sorted.bam | bcftools call -vmO z -o H3F3A.vcf.gz
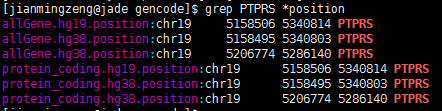
一个基因在同一套基因组上面竟然有两个定位!
查了好久的bug,终于搞清楚问题所在了!因为要对基因进行reads计数,所以要拿到基因在基因组上面的染色体起始终止坐标,结果发现了个十分诡异的现象,很多基因有多个坐标,比如下面这个PTPRS 在hg38这个基因组版本,居然有两个定位,因为我是写程序格式化得到的坐标,所以我check了我的程序,http://www.biotrainee.com/thread-472-1-1.html 感兴趣的同学可以点开看看我的代码!
mysql的table居然有最大列限制
【直播】我的基因组(五):测试数据及参考基因组的准备
我的全基因组数据还没拿到,而且还会推迟,简单说(tu)明(cao)一下原因(还好当初为了避免广告嫌疑一直没说是哪个公司负责测序,反正用的是illumina的hiseqX10这个测序啦,所以可以尽情的吐槽)。
心烦意乱的吐槽线 Continue reading
热图最佳实践-pheatmap
用pheatmap来绘图首先要安装这个包,它就一个功能,画出热图即可,号称是pretty heatmap,的确比其它的好用很多。我以前写过《一步一步学习heatmap.2》的教程,很简单的那种,所以就没有公布在博客上面,结果发现很多其它博客居然能先我一步发出。其实包括本次的pheatmap指南,都没什么好发,的在R里面也是傻瓜式出图,无法就是自己熟练一下参数而已,又不是开发一个包,没什么技术含量。我这里单独提一下pheatmap是因为它的确非常好用,将会是我画热图的不二之选。比如下面这个,是我最喜欢的: Continue reading
用samtools idxstats来对de novo的转录组数据计算表达量
de novo的转录组数据,比对的时候一般用的是自己组装好的trinity.fasta序列(挑选最长蛋白的转录本序列)来做参考,用bowtie2等工具直接将原始序列比对即可。所以比对 sam/bam文件本身就包含了参考序列的每一条转录本序列ID,直接对 sam/bam文件进行counts就知道每一个基因的表达量啦!
本来我是准备自己写脚本对sam文件进行counts就好,但是发现了samtools自带这样的工具:http://www.htslib.org/doc/samtools.html
如果是针对基因组序列,那么这个功能用处不大,但是针对转录本序列,统计出来的就是我们想要的转录本表达量。 Continue reading
【直播】我的基因组(四):计算资源的准备
大家久等了,Jimmy的测序数据还没有拿到手!但是,工欲善其事必先利其器!所以jimmy在等待自己基因组的这段时间里,准备好了自己计算资源!鉴于会有不少同志们会跟着直播来自己动手分析一个公共的全基因组重测序数据(参考数据下载地址详见直播一),我在这里和大家分享一下计算机资源及软件的基本要求。 Continue reading
【直播】我的基因组(三):抽血送样测序
【直播】我的基因组(二):科研和临床分析调研
那么我们一起来看看科研及临床一般都是进行怎样的分析吧! Continue reading