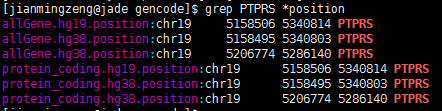
查了好久的bug,终于搞清楚问题所在了!因为要对基因进行reads计数,所以要拿到基因在基因组上面的染色体起始终止坐标,结果发现了个十分诡异的现象,很多基因有多个坐标,比如下面这个PTPRS 在hg38这个基因组版本,居然有两个定位,因为我是写程序格式化得到的坐标,所以我check了我的程序,http://www.biotrainee.com/thread-472-1-1.html 感兴趣的同学可以点开看看我的代码!
代码基本没有问题,我也去 genecard里面确认了PTPRS 的确只有一个坐标:http://www.genecards.org/cgi-bin/carddisp.pl?gene=PTPRS 那么为什么我的程序会得到两个不同的坐标呢?
我去搜索了该基因的记录,发现竟然有HAVANA和ENSEMBL的区别~~~
chr19 HAVANA gene 5158495 5340803 . - . gene_id "ENSG00000105426.15"; gene_type "protein_coding"; gene_status "KNOWN"; gene_name "PTPRS"; level 2; havana_gene "OTTHUMG00000180325.4";
chr19 ENSEMBL gene 5206774 5286140 . - . gene_id "ENSG00000283229.1"; gene_type "protein_coding"; gene_status "KNOWN"; gene_name "PTPRS"; level 3;
虽然我不知道什么意思,但是应该选择HAVANA才对!!!
For human, mouse, zebrafish, rat and pig, Ensembl not only shows transcripts that are annotated automatically using the Ensembl genebuild pipeline, but also transcripts that are manually annotated by the HAVANA team. If the Ensembl and Havana annotation agree with each other the transcripts are combined into an Ensembl/Havana merged transcript. When a transcript is only annotated by Ensembl or Havana it is named an Ensembl or Havana transcript, respectively. Transcripts that do match a species-specific entry in the UniProtKB/Swiss-Prot or RefSeq databases are categorised as known, those that do not as categorised as novel. For more detailed information, please have a look at our genebuild documentation.
而且根据这个可以看出,HAVANA 是一个验证团队,我们要相信他!!
还是太年轻呀,我以为选择了HAVANA就可以保证每个基因只有一个位置了,但是!
chr11 HAVANA gene 71505409 71529284 . - . gene_id "ENSG00000248671.7_2"; gene_type "processed_transcript"; gene_status "KNOWN"; gene_name "ALG1L9P"; level 2; tag "overlapping_locus"; havana_gene "OTTHUMG00000167480.2_2"; remap_status "full_contig"; remap_num_mappings 1; remap_target_status "overlap";
chr11 HAVANA gene 71511587 71515686 . - . gene_id "ENSG00000254978.2_1"; gene_type "transcribed_unprocessed_pseudogene"; gene_status "KNOWN"; gene_name "ALG1L9P"; level 2; tag "overlapping_locus"; havana_gene "OTTHUMG00000167481.1_1"; remap_status "full_contig"; remap_num_mappings 1; remap_target_status "overlap";
其实最主要的原因就是有多个ensembl数据库定义的基因都关联到同一个symbol,这个很麻烦,这个是Asparagine-Linked Glycosylation 1-Like 9, Pseudogene ,既然是Pseudogene,一般情况下的分析就应该过滤掉了算了!
但是也有protein coding的基因是有两个坐标的,我最后也是没有办法了,只好选择最长的基因咯
chr17 HAVANA gene 40177594 40250497 . - . gene_id "ENSG00000187595.15_2"; gene_type "protein_coding"; gene_status "KNOWN"; gene_name "ZNF385C"; level 1; tag "overlapping_locus"; havana_gene "OTTHUMG00000132073.6_2"; remap_status "full_contig"; remap_num_mappings 1; remap_target_status "overlap";
chr17 HAVANA gene 40190250 40202632 . - . gene_id "ENSG00000267221.2_2"; gene_type "protein_coding"; gene_status "KNOWN"; gene_name "ZNF385C"; level 2; tag "overlapping_locus"; havana_gene "OTTHUMG00000180103.2_2"; remap_status "full_contig"; remap_num_mappings 1; remap_target_status "overlap";