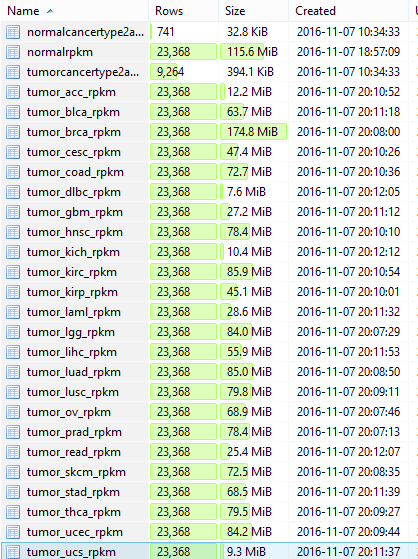
想着把TCGA的RPKM值矩阵表格写入到mysql,然后做一个查询网页给生物学家,我下载的是所有TCGA收集的mRNA表达数据集数据集-GSE62944 ,共9264个癌症样本,和741个正常组织的表达数据。当我想写入癌症表达矩阵的时候,报错了:
Error in .local(conn, statement, ...) :
could not run statement: Too many columns
简单搜索了一下,发现是mysql有最大列的限制,但是我不是很懂计算机,所以没太看明白该如何调整参数使得mysql列限制扩充:http://dev.mysql.com/doc/refman/5.7/en/column-count-limit.html 所以就把癌症表达矩阵根据癌症拆分了,癌种数量如下:
table(tumorCancerType2amples$CancerType)
ACC BLCA BRCA CESC COAD DLBC GBM HNSC KICH KIRC KIRP LAML LGG LIHC LUAD LUSC OV PRAD READ SKCM STAD THCA UCEC UCS
79 414 1119 306 483 48 170 504 66 542 291 178 532 374 541 502 430 502 167 472 420 513 554 57
分开写入mysql,下面给出解决方案及代码:
tumorRPKM=read.table('GSM1536837_06_01_15_TCGA_24.tumor_Rsubread_FPKM.txt.gz',sep = '\t',stringsAsFactors = F,header = T)colnames(tumorRPKM)[1]='geneSymbol'rownames(tumorRPKM)=tumorRPKM$geneSymboltumorRPKM=tumorRPKM[,-1]tumorRPKM=round( as.matrix(tumorRPKM),3)tumorRPKM=as.data.frame(tumorRPKM)tumorRPKM$geneSymbol = rownames(tumorRPKM)#load(file = 'tumorRPKM.rData')tumorCancerType2amples=read.table('GSE62944_06_01_15_TCGA_24_CancerType_Samples.txt',sep = '\t',stringsAsFactors = F)colnames(tumorCancerType2amples)=c('sampleID','CancerType')lapply(unique((tumorCancerType2amples$CancerType)), function(x){#x='PRAD';sampleList=tumorCancerType2amples[tumorCancerType2amples$CancerType==x,1]sampleList=gsub("-",".", sampleList)tmpMatrix=tumorRPKM[,c('geneSymbol',sampleList)]dbWriteTable(con, paste('tumor',x,'RPKM',sep='_'), tmpMatrix, append=F,row.names=F)})
dbWriteTable这个函数,需要加载RMySQL,而且还需要连接好mysql数据库,不然你根本就看不懂的!
写入数据库如下: