Monthly Archives: 1月 2016
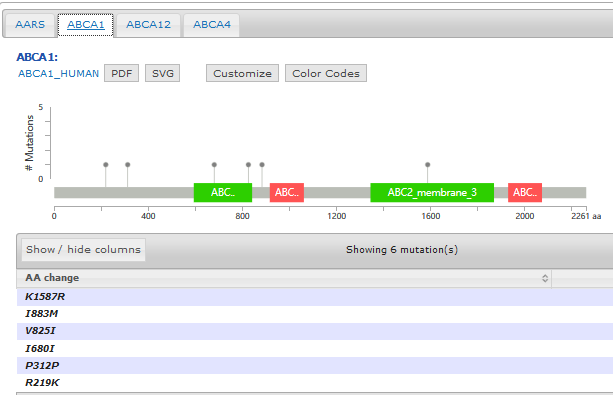
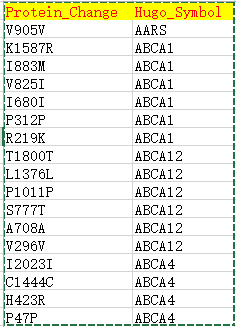
使用可视化工具MutationMapper来看看基因上面突变的分布
| Hugo_Symbol | HUGO symbol for the gene | TP53 |
| Protein_Change | Amino acid change | V600E |
- Support mutation data with annotated protein effects
- Mutation diagram/lollipop view
- Mutation table view
- 3D structure view if available
Pfam provides an online tool to not only generate the domain information in JSON format, but to draw the lollipop diagram using javascript as well. They have more information here: http://pfam.xfam.org/help#tabview=tab9
IMHO, not as pretty as cBioPortal's but it gets you close to a solution.
EDIT / SHAMELESS PLUG: After seeing the data available and how easy it'd be, I made my own quick tool to fetch the data and draw the diagram for me in a style similar to cBioPortal - feel free to fork it and add features: https://github.com/pbnjay/lollipops
Example output (w/ labels per the comments)
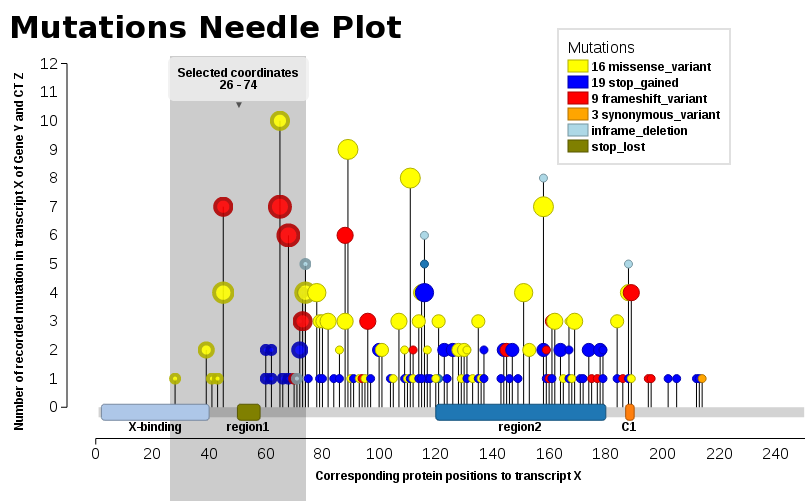
We found ourselves in the same need, we wanted such a plot (JavaScript). Thus, I add our solution, Mutations Needle Plot. The library creates an SVG image (with D3), which then may be downloaded.
- Live examples are found at BioJS: http://registry.biojs.net/client/#/detail/muts-needle-plot
- Code is available at GitHub: https://github.com/bbglab/muts-needle-plot
- And it is also a npm-package: https://www.npmjs.com/package/muts-needle-plot
You will npm in order to be able to install & run the library.
Examples may be found in the snippets folder or also the index.html - The one displayed here below
gene的symbol与entrez ID并不是绝对的一一对应的
很多时候,我们都无法确定到底是基于symbol来进行分析,还是基于entrez ID,当我们要进行ID转换的时候也想当然的以为它们的一一对应的, 但是最近我写了一个脚本来分析CCLE的数据的时候,发现其实有一些特例:
all_symbol=mappedkeys(org.Hs.egSYMBOL2EG)
用RankComp的思想来做差异基因分析
我现在还不是很确定这个方法,只是试一试,欢迎与我交流对该方法的讨论!
Wang H, Sun Q, Zhao W, et al. Individual-level analysis of differential expression of genes and pathways for personalized medicine[J]. Bioinformatics, 2014: btu522.
他们把它写成了一个R包,可以下载使用,但是必须用R2.15.2版本,我用了一下,不好用!
We can download the R code for in http://bioinformatics.oxfordjournals.org/content/31/1/62/suppl/DC1
他们这个程序真心不好用,但是很容易看懂算法,可以自己用R语言写一个来实现同样的过程!
用TCGA数据做cox生存分析的风险因子(比例风险模型)
用my.surv <- surv(OS_MONTHS,OS_STATUS=='DECEASED')构建生存曲线。用kmfit2 <- survfit(my.surv~TUMOR_STAGE_2009)来做某一个因子的KM生存曲线。用 survdiff(my.surv~type, data=dat)来看看这个因子的不同水平是否有显著差异,其中默认用是的logrank test 方法。用coxph(Surv(time, status) ~ ph.ecog + tt(age), data=lung) 来检测自己感兴趣的因子是否受其它因子(age,gender等等)的影响。
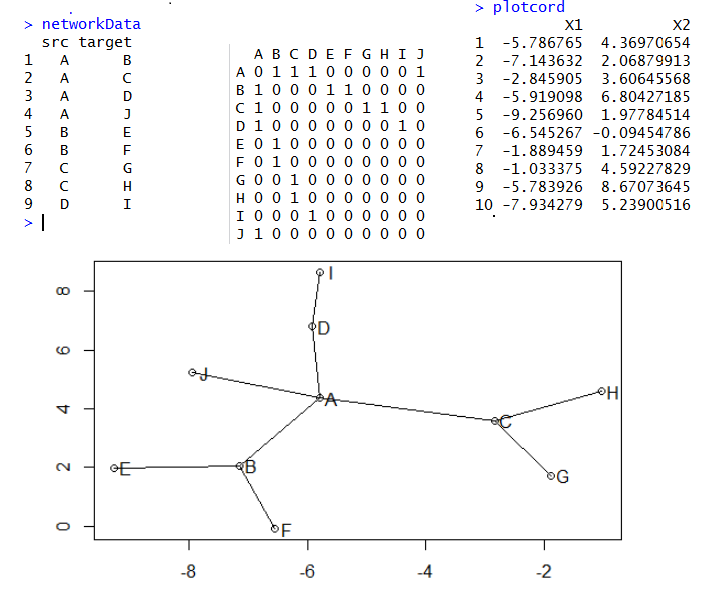
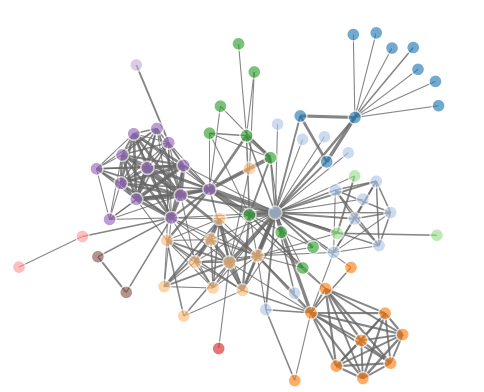
R语言画网络图三部曲之sna
如果只是画网络图,那么只需要把所有的点,按照算好的坐标画出来,然后把所有的连线也画出即可!
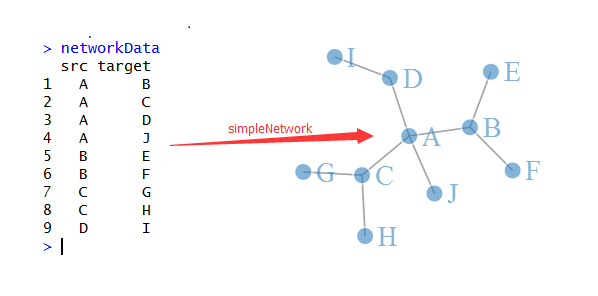
R语言画网络图三部曲之networkD3
接下来, 我们直接看看R里面是如何画网络图的,我们首推一个包:networkD3/
用broad出品的软件来处理bam文件几次遇到文件头错误
报错如下:ERROR MESSAGE: SAM/BAM file input.marked.bam is malformed: SAM file doesn't have any read groups defined in the header. The GATK no longer supports SAM files without read groups !
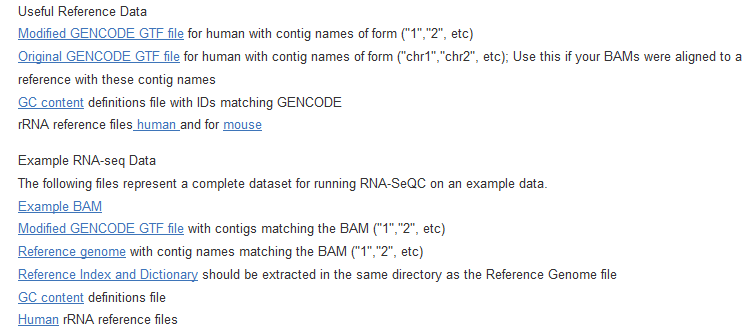
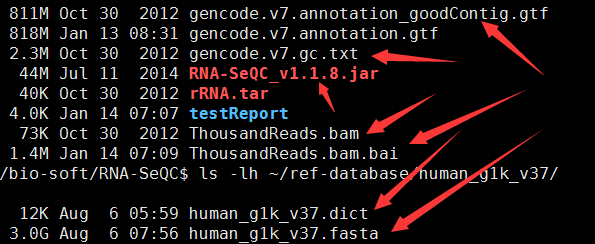
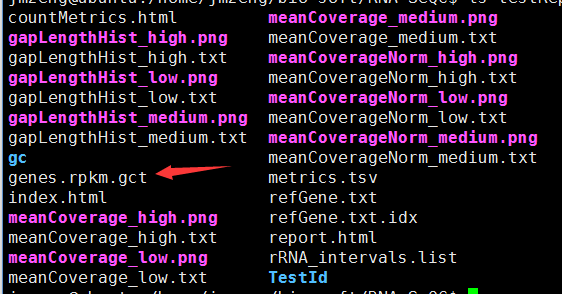
用RNA-SeQC得到表达矩阵RPKM值
这个软件不仅仅能做QC,而且可以统计各个基因的RPKM值!尤其是TCGA计划里面的都是用它算的
一、程序安装
java -jar RNASeQC.jar -n 1000 -s "TestId|ThousandReads.bam|TestDesc" -t gencode.v7.annotation_goodContig.gtf -r Homo_sapiens_assembly19.fasta -o ./testReport/ -strat gc -gc gencode.v7.gc.txt
-n 1000 \
-s "TestId|ThousandReads.bam|TestDesc" \
-t gencode.v7.annotation_goodContig.gtf \
-r ~/ref-database/human_g1k_v37/human_g1k_v37.fasta \
-o ./testReport/ \
-strat gc \
-gc gencode.v7.gc.txt \
java -jar RNASeQC.jar -n 1000 -s "TestId|ThousandReads.bam|TestDesc" -t gencode.v7.annotation_goodContig.gtf -r Homo_sapiens_assembly19.fasta -o ./testReport/ -strat gc -gc gencode.v7.gc.txt -BWArRNA human_all_rRNA.fasta
Note: this assumes BWA is in your PATH. If this is not the case, use the -bwa flag to specify the path to BWA
TCGA数据里面都会提供由RNA-SeQC软件处理得到的表达矩阵!
Expression
- RPKM data are used as produced by RNA-SeQC.
- Filter on >=10 individuals with >0.1 RPKM and raw read counts greater than 6.
- Quantile normalization was performed within each tissue to bring the expression profile of each sample onto the same scale.
- To protect from outliers, inverse quantile normalization was performed for each gene, mapping each set of expression values to a standard normal.
华盛顿大学把所有的变异数据都用自己的方法注释了一遍,然后提供下载
A general framework for estimating the relative pathogenicity of human genetic variants.
Nat Genet. 2014 Feb 2. doi: 10.1038/ng.2892.
PubMed PMID: 24487276.
蛋白质相互作用(PPI)数据库大全
Your search returned 207 results in 9 categories with the following search parameters:
- Organisms: Homo sapiens (Human)
- Availability: Free to all users
- Standards: all
| BIND | the biomolecular interaction network database | died link |
| DIP | the database of interacting proteins | http://dip.doe-mbi.ucla.edu/ |
| MINT | the molecular interaction database | http://mint.bio.uniroma2.it/mint/ |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins | http://string-db.org/ |
| HPRO | Human protein reference database | http://www.hprd.org/ |
| BioGRID | The Biological General Repository for Interaction Datasets | http://thebiogrid.org/ |
居然可以下载千人基因组计划的所有数据bam,vcf数据
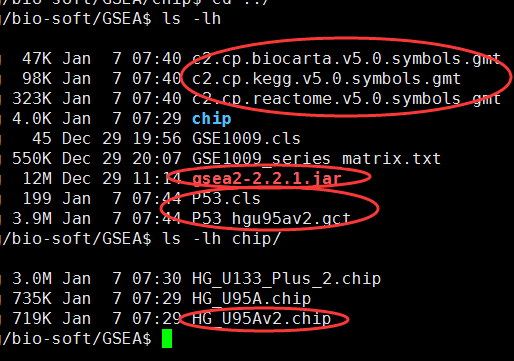
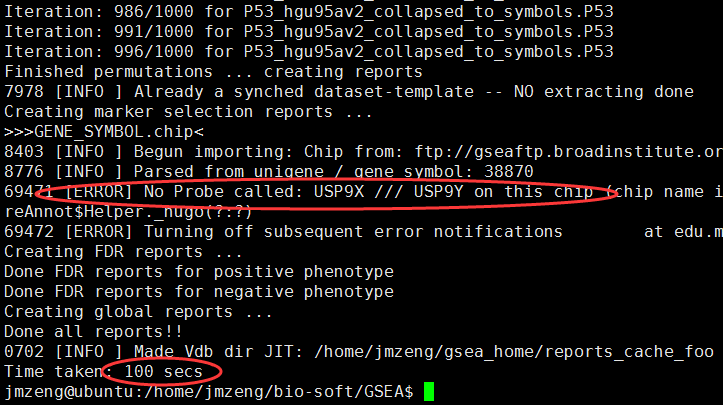
批量运行GSEA,命令行版本
之前用过有界面的那种,那样非常方便,只需要做好数据即可,但是如果有非常多的数据,每次都要点击文件,点击下一步,也很烦,不过,,它既然是java软件,就可以以命令行的形式来玩转它!
直接在官网下载java版本软件即可:http://software.broadinstitute.org/gsea/downloads.jsp
需要下载gmt文件,自己制作gct和cls文件,或者直接下载测试文件p53
hgu95av2的芯片数据,只有一万多探针,所以很快就可以出结果
| CP: Canonical pathways (browse 1330 gene sets) |
Gene sets from the pathway databases. Usually, these gene sets are canonical representations of a biological process compiled by domain experts. details | Download GMT Files original identifiers gene symbols entrez genes ids |
|---|---|---|
| CP:BIOCARTA: BioCarta gene sets (browse 217 gene sets) |
Gene sets derived from the BioCarta pathway database (http://www.biocarta.com/genes/index.asp). | Download GMT Files original identifiers gene symbols entrez genes ids |
| CP:KEGG: KEGG gene sets (browse 186 gene sets) |
Gene sets derived from the KEGG pathway database (http://www.genome.jp/kegg/pathway.html). | Download GMT Files original identifiers gene symbols entrez genes ids |
| CP:REACTOME: Reactome gene sets (browse 674 gene sets) |
Gene sets derived from the Reactome pathway database (http://www.reactome.org/). | Download GMT Files original identifiers gene symbols entrez genes ids |
关于芯片平台GPL15308和GPL570
它们虽然被GEO数据标记了不同的ID号,但是其实都是一种芯片,都是昂飞公司的U133++2芯片,分析过芯片数据的人肯定不会陌生了
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL15308
事实上,这个平台应该是GPL570,但是被CCLE数据库给稍微变通了一下,就给了一个GPL15308的标签,平台主页也写的很清楚,它的探针ID是伪ID,其实就是entrez gene ID
| 本来这个芯片设计的是五万多个探针,最后只剩下了18926个基因 |
| This array is identical to GPL570 but the data were analyzed with a custom CDF Brainarray Version 15, hgu133plus2hsentrezg. |
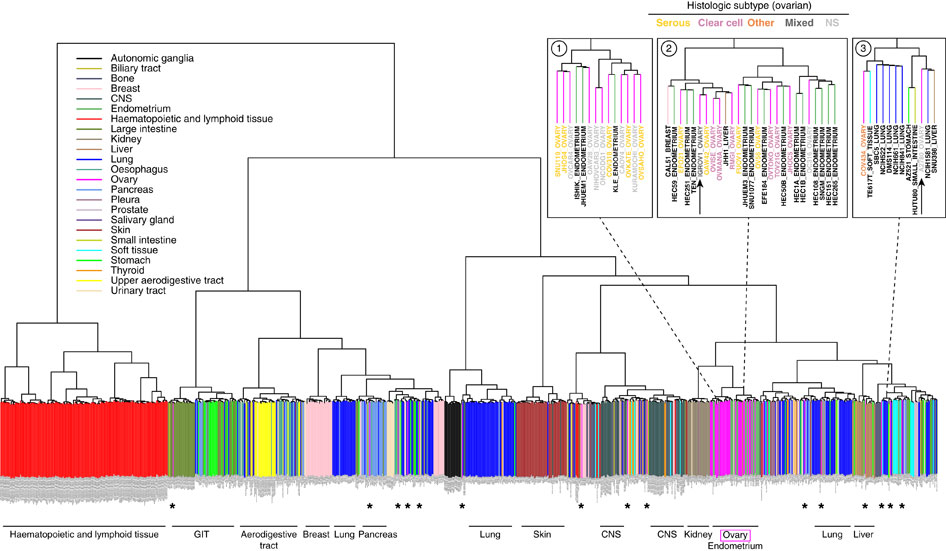
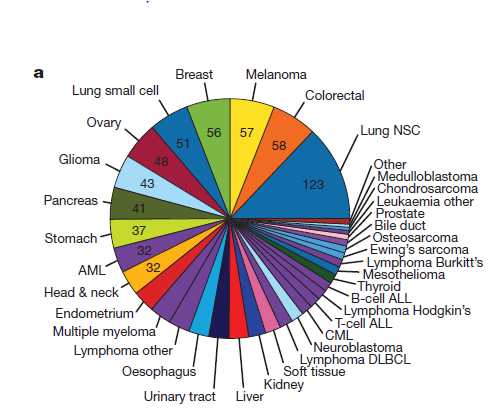
对CCLE数据库可以做的分析
step4:Unsupervised hierarchical clustering (1-Spearman distance, average linkage) was performed on the cell lines using the aCGH data.
Putative driver genes of which copy number aberrations correlated to mRNA gene expression were identified to determine subtypes or clusters that are driven by different mechanisms. This was done using Mann Whitney U-test with p<0.05, and Spearman Correlation Coefficient test with Rho >0.6.
step5:We then performed consensus clustering[17] on the gene expression data of the 27 gastric cancer cell lines from CCLE using these putative driver genes. We selected k = 2 as it gives sufficiently stable similarity matrix.
step6: In order to assign new samples to this integrative cluster, significance analysis of microarray (SAM) [18]with threshold q<2.0 was used to generate subtype signature based on the mRNA expression data of the 1762 genes from the 27 gastric cancer cell lines in CCLE.
先用甲基化数据来聚类,得到putative driver genes,然后再用这些基因的表达数据来再次聚类,分成两类,然后对这两类进行SAM找差异基因
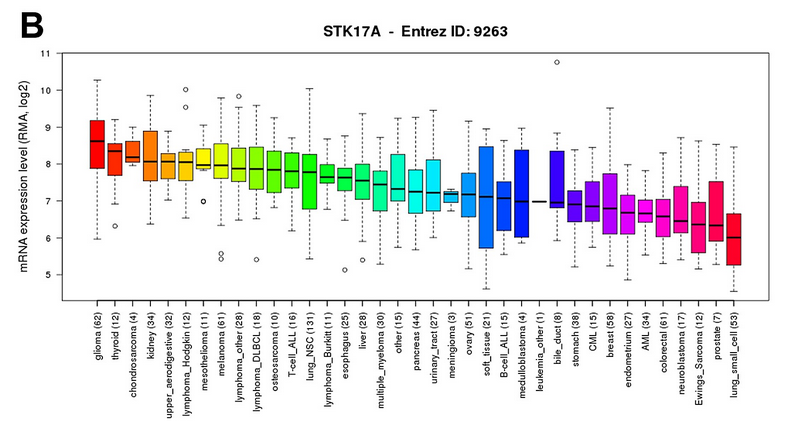
结论就是:STK17A is highly expressed in glioma cell lines compared to other cancer types. Data was obtained through the Cancer Cell Line Encyclopedia (CCLE).
第三篇文献:http://www.nature.com/ncomms/2013/130709/ncomms3126/fig_tab/ncomms3126_F4.html
CCLE数据库几个知识点
TCGA数据里面的生存分析例子
生存分析简介
survfit:创建KM生存曲线或是Cox调整生存曲线
survdiff:用于不同组的统计检验
别人写的代码运行真快!!!
源码在这个package的github里面可以找到,有兴趣的童鞋可以研究一下
R语言软件的各种旧版本下载
然后我看了一下域名,发现其实很有规律的
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
linux/ | 23-Jan-2008 19:47 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
macos/ | 19-Apr-2005 09:45 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
macosx/ | 12-Dec-2015 09:04 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
windows/ | 24-Feb-2012 18:41 | - |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
debian/ | 15-Dec-2015 02:06 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
redhat/ | 27-Jul-2014 21:12 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
suse/ | 16-Feb-2012 15:09 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
ubuntu/ | 06-Jan-2016 04:05 | - |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
precise/ | 06-Jan-2016 04:03 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
trusty/ | 06-Jan-2016 04:04 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
vivid/ | 06-Jan-2016 04:04 | - | |
![[DIR]](file:///C:/Users/jmzeng/AppData/Local/YNote/data/jmzeng1314@163.com/901f79782af94aecb22c22dcb42c10a4/folder.gif) |
wily/ | 06-Jan-2016 04:05 | - |
所以如果,大家是想在linux系统里面安装旧版本的R,建议大家直接下载c源码,直接三部曲就可以安装啦!
R 3.2.1 (June, 2015)
R 3.2.0 (April, 2015)
R 3.1.3 (March, 2015)
R 3.1.2 (October, 2014)
R 3.1.1 (July, 2014)
R 3.1.0 (April, 2014)
R 3.0.3 (March, 2014)
R 3.0.2 (September, 2013)
R 3.0.1 (May, 2013)
R 3.0.0 (April, 2013)
R 2.15.3 (March, 2013)
R 2.15.2 (October, 2012)
R 2.15.1 (June, 2012)
R 2.15.0 (March, 2012)
R 2.14.2 (February, 2012)
R 2.14.1 (December, 2011)
R 2.14.0 (November, 2011)
R 2.13.2 (September, 2011)
R 2.13.1 (July, 2011)
R 2.13.0 (April, 2011)
R 2.12.2 (February, 2011)
R 2.12.1 (December, 2010)
R 2.12.0 (October, 2010)
R 2.11.1 (May, 2010)
R 2.11.0 (April, 2010)
R 2.10.1 (December, 2009)
R 2.10.0 (October, 2009)
R 2.9.2 (August, 2009)
R 2.9.1 (June, 2009)
R 2.9.0 (April, 2009)
R 2.8.1 (December, 2008)
R 2.8.0 (October, 2008)
R 2.7.2 (August, 2008)
R 2.7.1 (June, 2008)
R 2.7.0 (April, 2008)
R 2.6.2 (February, 2008)
R 2.6.1 (November, 2007)
R 2.6.0 (October, 2007)
R 2.5.1 (July, 2007)
R 2.5.0 (April, 2007)
R 2.4.1 (December, 2006)
R 2.4.0 (October, 2006)
R 2.3.1 (June, 2006)
R 2.3.0 (April, 2006)
R 2.2.1 (December, 2005)
R 2.2.0 (October, 2005)
R 2.1.1 (June, 2005)
R 2.1.0 (April, 2005)
R 2.0.1 (November, 2004)
R 2.0.0 (October, 2004)
R 1.9.1 (June, 2004)
R 1.8.1 (November, 2003)
R 1.7.1 (June, 2003)
R 1.6.2 (January, 2003)
Installer for R 1.5.1 (June, 2002)
Installer for R 1.4.1 (January, 2002)
Installer for R 1.3.1 (September, 2001)
Binary files for R 1.2.2 (March, 2001)
Binary files for R 1.0.0 (February, 2000)