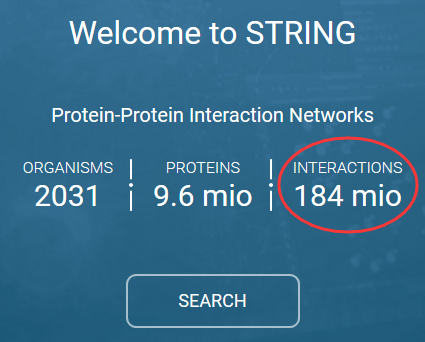
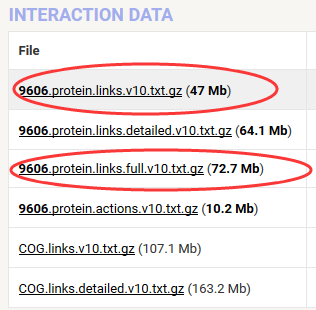
一个比较综合的链接是:A compendium of PPI databases can be found in
http://www.pathguide.org/.
里面的数据库非常多,仅仅是对于人类就有
Your search returned 207 results in 9 categories with the following search parameters:
人类的六个主要PPI是:Analysis of human interactome PPI data showing the coverage of six major primary databases (BIND, BioGRID, DIP, HPRD, IntAct, and MINT), according to the integration provided by the meta-database APID.
这些数据库大部分都还有维护者,还在持续更新,每次更新都可以发一篇paper,而数据库收集的paper引用一般都上千,如果你做了一个数据库,才十几个人引用,那就说明你是自己在跟自己玩。
(a) PPI definition; a definition of a protein-to-protein interaction compared to other biomolecular relationships or associations.
(b)PPI determination by two alternative approaches: binary and co-complex; a description of the PPIs determined by the two main types of experimental technologies.
(c) The main databases and repositories that include PPIs; a description and comparison of the main databases and repositories that include PPIs, indicating the type of data that they collect with a special distinction between experimental and predicted data.
(d) Analysis of coverage and ways to improve PPI reliability; a comparative study of the current coverage on PPIs and presentation of some strategies to improve the reliability of PPI data.
(e) Networks derived from PPIs compared to canonical pathways; a practical example that compares the characteristics and information provided by a canonical pathway and the PPI network built for the same proteins. Last, a short summary and guidance for learning more is provided.
现在的蛋白质相互作用数据库的数据都很有限,但是在持续增长,一般有下面四种原因导致数据被收录到数据库
There are four common approaches for PPI data expansions:
1) manual curation from the biomedical literature by experts;
2) automated PPI data extraction from biomedical literature with text mining methods;
3) computational inference based on interacting protein domains or co-regulation relationships, often derived from data in model organisms; and
4) data integration from various experimental or computational sources.
Partly due to the difficulty of evaluating qualities for PPI data, a majority of widely-used PPI databases, including DIP, BIND, MINT, HPRD, and IntAct, take a "conservative approach" to PPI data expansion by adding only manually curated interactions. Therefore, the coverage of the protein interactome developed using this approach is poor.
In the second literature mining approach, computer software replaces database curators to extract protein interaction (or, association) data from large volumes of biomedical literature . Due to the complexity of natural language processing techniques involved, however, this approach often generates large amount of false positive protein "associations" that are not truly biologically significant "interactions".
The challenge for the integrative approach is how to balance quality with coverage.
In particular, different databases may contain many redundant PPI information derived from the same sources, while the overlaps between independently derived PPI data sets are quite low .
参考: