Monthly Archives: 2月 2016
在R里面操作SQLite
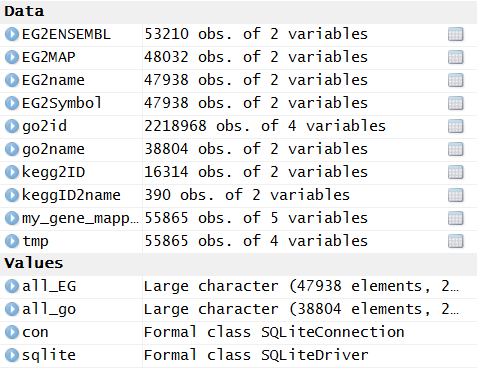
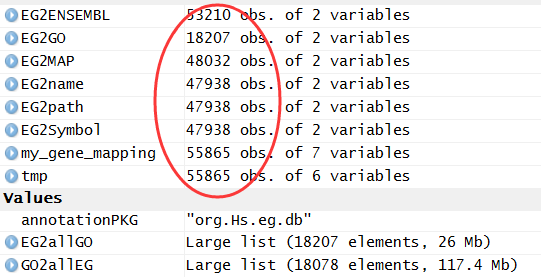
library(org.Hs.eg.db )
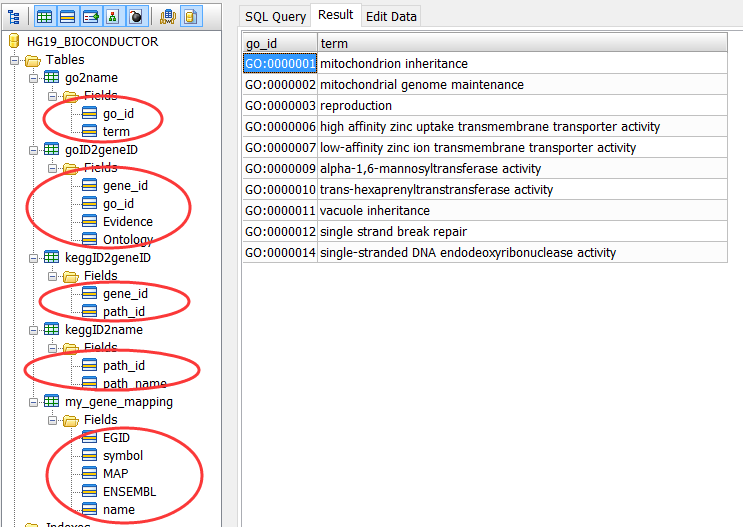
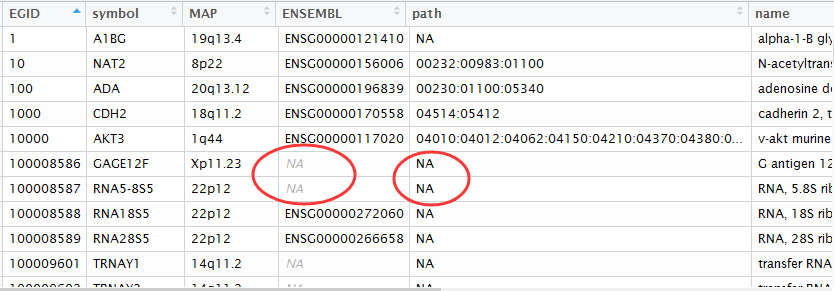
把bioconductor的gene mapping信息上传到mysql数据库
用crossmap代替liftover做基因组坐标转换
This online tool currently uses CrossMap, which supports a limited number of formats (see our online documentation for details of the individual data formats listed below). CrossMap also discards metadata in files, so track definitions, etc, will be lost on conversion.
Important note: CrossMap converts WIG files to BedGraph internally for efficiency, and also outputs them in BedGraph format.
没必要学shell进阶语法
因为大部分生物信息学软件都是linux版本的,所以生物信息学数据分析工作者必备技能就是linux,但是大部分人只是拿他当个中转站,我以前也是,直到接触了大批量的任务,自动化流程,才明白这里面的水太深了,不过无所谓,凭我个人的观点,其实shell的进阶语法真的不必要!
我也不想去搞明白操作符两边是否加空格的区别是什么了。
如果,还有其它功能需要实现,我们可以把脚本写的更负载,纯粹的用shell,需要搜索更多的shell技巧。
[perl]
## perl nohup.pl deep_count.sh 0
## perl nohup.pl deep_count.sh 1
## perl nohup.pl deep_count.sh 2
$i=1;
open FH,$ARGV[0];
while(<FH>){
chomp;
next unless $.%3==$ARGV[1];
$cmd="nohup $_ &";
print "$cmd\n";
system($cmd);
sleep(10800) if $i%5==4;
$i++;
#exit;
}
[/perl]
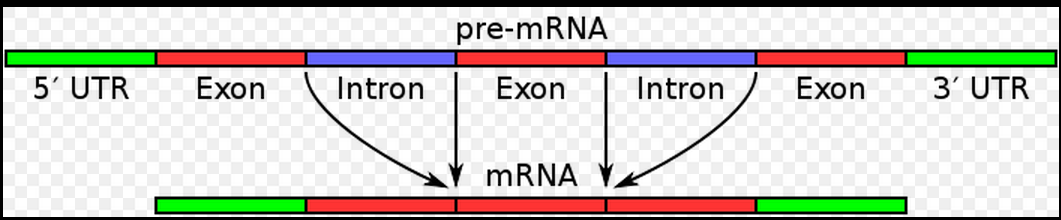
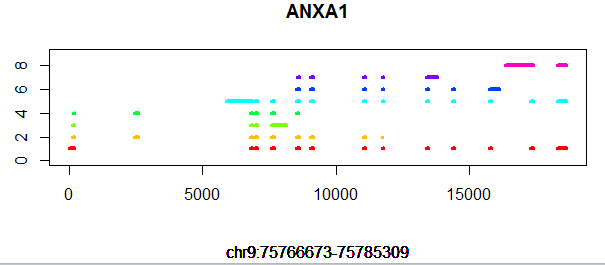
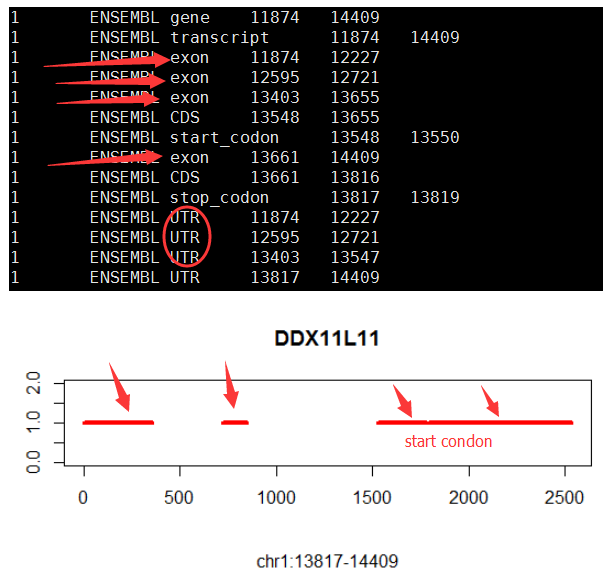
画基因结构图!
[perl]
suppressMessages(library(ggplot2))
suppressMessages(library(RMySQL))
con <- dbConnect(MySQL(), host="127.0.0.1", port=3306, user="root", password="11111111")
dbSendQuery(con, "USE test")
gene='SOX10'
#gene='DDX11L11'
if (T){
query=paste("select * from hg19_gtf where gene_type='protein_coding' and gene_name=",shQuote(gene),sep="")
structure=dbGetQuery(con,query)
tmp_min=min(c(structure$start,structure$end))
structure$new_start=structure$start-tmp_min
structure$new_end=structure$end-tmp_min
tmp_max=max(c(structure$new_start,structure$new_end))
num_transcripts=nrow(structure[structure$record=='transcript',])
tmp_color=rainbow(num_transcripts)
x=1:tmp_max;y=rep(num_transcripts,length(x))
#x=10000:17000;y=rep(num_transcripts,length(x))
plot(x,y,type = 'n',xlab='',ylab = '',ylim = c(0,num_transcripts+1))
title(main = gene,sub = paste("chr",tmp$chr,":",tmp$start,"-",tmp$end,sep=""))
j=0;
tmp_legend=c()
for (i in 1:nrow(structure)){
tmp=structure[i,]
if(tmp$record == 'transcript'){
j=j+1
tmp_legend=c(tmp_legend,paste("chr",tmp$chr,":",tmp$start,"-",tmp$end,sep=""))
}
if(tmp$record == 'exon') lines(c(tmp$new_start,tmp$new_end),c(j,j),col=tmp_color[j],lwd=4)
}
# legend('topleft',legend=tmp_legend,lty=1,lwd = 4,col = tmp_color);
}
[/perl]
生物信息学分析过程中常见文件格式
刚开始接触生物信息学的时候我也很纠结什么fastq,fastq,sam,bam,vcf,maf,gtf,bed,psl等等,甚至还有过时了的NCBI,ENSEMBL格式,如果是我刚开始 学的时候,我倒是很愿意把他们全部搞透彻,写详细的说明书,但是现在成长了,这些东西感觉很low了,正好我看到了一篇帖子讲数据格式的收集大全,分享给大家,希望初学者能多花点时间好好钻研!
https://www.biostars.org/p/55351/
用R获取芯片探针与基因的对应关系三部曲-bioconductor
现有的基因芯片种类不要太多了!
gpl organism bioc_package1 GPL32 Mus musculus mgu74a2 GPL33 Mus musculus mgu74b3 GPL34 Mus musculus mgu74c6 GPL74 Homo sapiens hcg1107 GPL75 Mus musculus mu11ksuba8 GPL76 Mus musculus mu11ksubb9 GPL77 Mus musculus mu19ksuba10 GPL78 Mus musculus mu19ksubb11 GPL79 Mus musculus mu19ksubc12 GPL80 Homo sapiens hu680013 GPL81 Mus musculus mgu74av214 GPL82 Mus musculus mgu74bv215 GPL83 Mus musculus mgu74cv216 GPL85 Rattus norvegicus rgu34a17 GPL86 Rattus norvegicus rgu34b18 GPL87 Rattus norvegicus rgu34c19 GPL88 Rattus norvegicus rnu3420 GPL89 Rattus norvegicus rtu3422 GPL91 Homo sapiens hgu95av223 GPL92 Homo sapiens hgu95b24 GPL93 Homo sapiens hgu95c25 GPL94 Homo sapiens hgu95d26 GPL95 Homo sapiens hgu95e27 GPL96 Homo sapiens hgu133a28 GPL97 Homo sapiens hgu133b29 GPL98 Homo sapiens hu35ksuba30 GPL99 Homo sapiens hu35ksubb31 GPL100 Homo sapiens hu35ksubc32 GPL101 Homo sapiens hu35ksubd36 GPL201 Homo sapiens hgfocus37 GPL339 Mus musculus moe430a38 GPL340 Mus musculus mouse430239 GPL341 Rattus norvegicus rae230a40 GPL342 Rattus norvegicus rae230b41 GPL570 Homo sapiens hgu133plus242 GPL571 Homo sapiens hgu133a243 GPL886 Homo sapiens hgug4111a44 GPL887 Homo sapiens hgug4110b45 GPL1261 Mus musculus mouse430a249 GPL1352 Homo sapiens u133x3p50 GPL1355 Rattus norvegicus rat230251 GPL1708 Homo sapiens hgug4112a54 GPL2891 Homo sapiens h20kcod55 GPL2898 Rattus norvegicus adme16cod60 GPL3921 Homo sapiens hthgu133a63 GPL4191 Homo sapiens h10kcod64 GPL5689 Homo sapiens hgug4100a65 GPL6097 Homo sapiens illuminaHumanv166 GPL6102 Homo sapiens illuminaHumanv267 GPL6244 Homo sapiens hugene10sttranscriptcluster68 GPL6947 Homo sapiens illuminaHumanv369 GPL8300 Homo sapiens hgu95av270 GPL8490 Homo sapiens IlluminaHumanMethylation27k71 GPL10558 Homo sapiens illuminaHumanv472 GPL11532 Homo sapiens hugene11sttranscriptcluster73 GPL13497 Homo sapiens HsAgilentDesign02665274 GPL13534 Homo sapiens IlluminaHumanMethylation450k75 GPL13667 Homo sapiens hgu21976 GPL15380 Homo sapiens GGHumanMethCancerPanelv177 GPL15396 Homo sapiens hthgu133b78 GPL17897 Homo sapiens hthgu133a
gpl_info=read.csv("GPL_info.csv",stringsAsFactors = F)### first download all of the annotation packages from bioconductorfor (i in 1:nrow(gpl_info)){print(i)platform=gpl_info[i,4]platform=gsub('^ ',"",platform) ##主要是因为我处理包的字符串前面有空格#platformDB='hgu95av2.db'platformDB=paste(platform,".db",sep="")if( platformDB %in% rownames(installed.packages()) == FALSE) {BiocInstaller::biocLite(platformDB)#source("http://bioconductor.org/biocLite.R");#biocLite(platformDB )}}
下载完了所有的包, 就可以进行批量导出芯片探针与gene的对应关系!
for (i in 1:nrow(gpl_info)){print(i)platform=gpl_info[i,4]platform=gsub('^ ',"",platform)#platformDB='hgu95av2.db'platformDB=paste(platform,".db",sep="")if( platformDB %in% rownames(installed.packages()) != FALSE) {library(platformDB,character.only = T)#tmp=paste('head(mappedkeys(',platform,'ENTREZID))',sep='')#eval(parse(text = tmp))###重点在这里,把字符串当做命令运行all_probe=eval(parse(text = paste('mappedkeys(',platform,'ENTREZID)',sep='')))EGID <- as.numeric(lookUp(all_probe, platformDB, "ENTREZID"))##自己把内容写出来即可}}