前面我带领大家通过IMGT数据库认知免疫组库,而且也一起从IMGT数据库下载免疫组库相关fasta序列,免疫组库重要的研究对象就是分成BCR的IGH,IGK,IGL这3类,以及TCR的TRA,TRB,TRD,TRG,它们各自都有V,D(可选),J,C基因。
接下来又认识了免疫组库测序数据,知道了免疫组库测序数据的一些特性,并且使用igblast进行免疫组库分析了,但是那个是初步的比对,虽然找到每一个测序片段的V,D(可选),J,C基因,并且拿到 CDR3序列,中间步骤有点多,常规的测序数据过滤就算了,还需要把PE数据合并,fastq格式转为fasta格式,而且本身igblast软件就很难使用,数据库文件构建也繁琐。其实也有包装好的一站式流程。
免疫组库测序数据可以使用MiXCR进行分析,比如文章 T cell receptor next-generation sequencing reveals cancer-associated repertoire metrics and reconstitution after chemotherapy in patients with hematological and solid tumors ,就提到了:
NGS and de-multiplexing was performed on an Illumina MiSeq sequencer (600–cycle, single indexed, paired-end run). Analysis of the TRB locus was computed using the MiXCR analysis tool
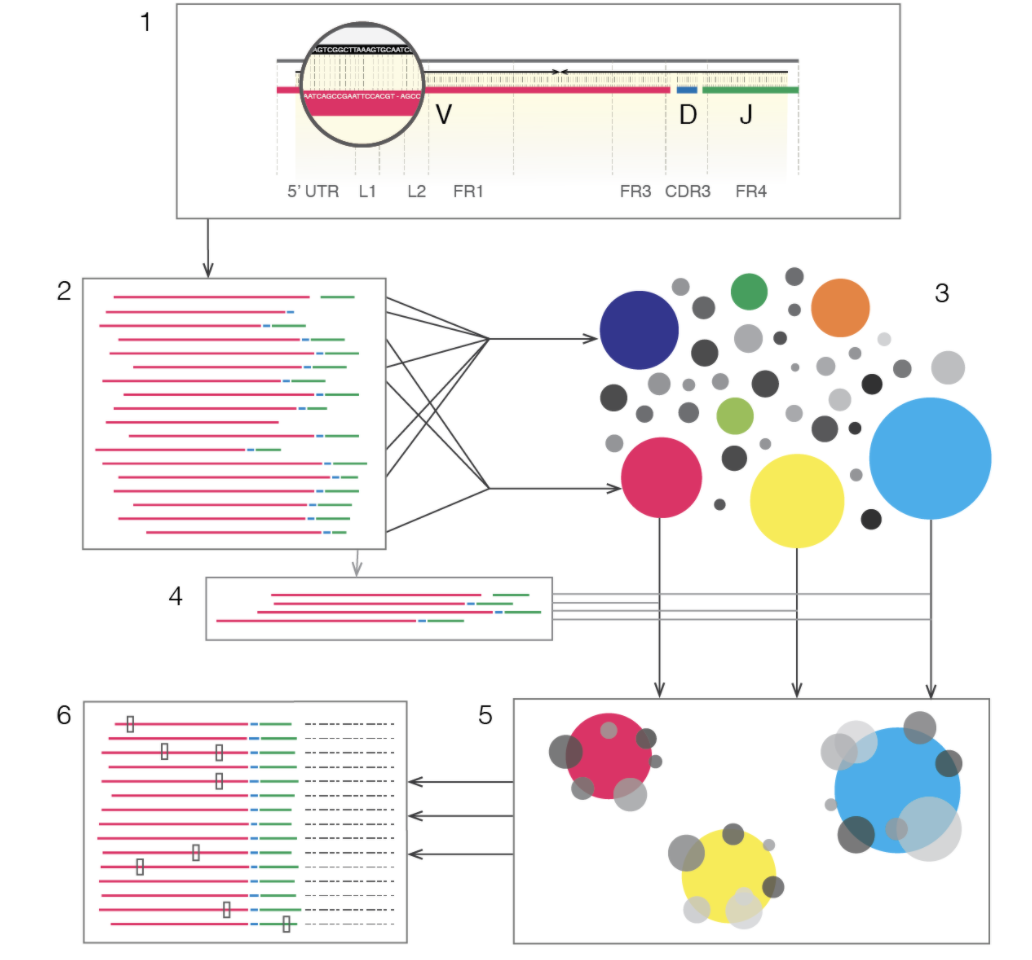
MiXCR是MiLaboratory开发的,他们实验室出品了3款:MiXCR, MIGEC, VDJtools ,在免疫组库领域都很出名。其中MiXCR工作流程示意图如下:
安装MiXCR软件
因为MiXCR有二进制版本,所以推荐直接下载它并且解压即可使用啦!
- https://github.com/milaboratory/mixcr/releases/download/v3.0.13/mixcr-3.0.13.zip (2020年05月05日查看的最新版)
- 文档:https://mixcr.readthedocs.io/en/latest/index.html
不过MiXCR是基于java平台开发,所以启动MiXCR时候,需要保证java环境是ok的,而且要求java8哦。
如果是Linux或者macOS,那么非常简单,直接使用里面的mixcr命令即可调用该软件。
如果是Windows平台呢,需要使用 java -Xmx4g -Xms3g -jar 来调用下载到的MiXCR有二进制版本里面的jar文件哦。
我的是macOS操作系统,软件下载解压后如下所示:
(base) jmzengdeMBP:mixcr-3.0.13 jmzeng$ pwd
/Users/jmzeng/biosoft/MiXCR/mixcr-3.0.13
(base) jmzengdeMBP:mixcr-3.0.13 jmzeng$ tree -h
.
├── [1.4K] LICENSE
├── [ 96] libraries
├── [4.0K] mixcr
└── [9.3M] mixcr.jar
1 directory, 3 files
一站式流程使用
MiXCR软件提供一个 analyze模式,可以一站式完成(align, assemblePartial, extend, assemble, assembleContigs and export) 这些分析,需要认真读 https://mixcr.readthedocs.io/en/latest/analyze.html 说明书,其提供的例子如下:
mixcr analyze amplicon
-s <species> \
--starting-material <startingMaterial> \
--5-end <5End> --3-end <3End> \
--adapters <adapters> \
[OPTIONS] input_file1 [input_file2] analysis_name
在前面我们认识的免疫组库测序数据,是人类的,MiSeq测序仪,PE300测序策略,TRB,DNA测序。所以我们构建的命令是:
mixcr=/Users/jmzeng/biosoft/MiXCR/mixcr-3.0.13/mixcr
$mixcr analyze amplicon \
-s hsa \
--starting-material dna \
--5-end v-primers --3-end c-primers \
--adapters adapters-present \
raw/ERR3445170_1.fastq.gz raw/ERR3445170_2.fastq.gz ERR3445170
因为我设置了 —adapters adapters-present,所以这个软件也会帮忙去除我们测序数据里面的接头序列,所以我这里直接使用了raw测序数据fq文件进行分析,无需经过前面igblastn的trim_galore过程。
很轻松就出来了结果,看起来调用了我Mac全部计算资源
运行日志如下;
NOTE: report file is not specified, using ERR3445170.report to write report.
Alignment: 2.1%
Alignment: 15.4% ETA: 00:00:38
Alignment: 27.9% ETA: 00:00:17
Alignment: 39.5% ETA: 00:00:15
Alignment: 51.5% ETA: 00:00:12
Alignment: 63% ETA: 00:00:09
Alignment: 74.2% ETA: 00:00:06
Alignment: 85.4% ETA: 00:00:03
Alignment: 96.9% ETA: 00:00:00
============= Report ==============
Analysis time: 29.52s
Total sequencing reads: 84833
Successfully aligned reads: 83421 (98.34%)
Paired-end alignment conflicts eliminated: 3027 (3.57%)
Alignment failed, no hits (not TCR/IG?): 850 (1%)
Alignment failed because of absence of V hits: 4 (0%)
Alignment failed because of absence of J hits: 507 (0.6%)
No target with both V and J alignments: 49 (0.06%)
Alignment failed because of low total score: 2 (0%)
Overlapped: 81097 (95.6%)
Overlapped and aligned: 80179 (94.51%)
Alignment-aided overlaps: 1313 (1.64%)
Overlapped and not aligned: 918 (1.08%)
V gene chimeras: 29 (0.03%)
J gene chimeras: 4 (0%)
TRB chains: 83418 (100%)
IGH chains: 3 (0%)
Realigned with forced non-floating bound: 10098 (11.9%)
Realigned with forced non-floating right bound in left read: 3 (0%)
Realigned with forced non-floating left bound in right read: 3 (0%)
Initialization: progress unknown
Assembling initial clonotypes: 100%
Building clones: 25.2%
Writing clones: 0%
============= Report ==============
Analysis time: 2.33s
Final clonotype count: 2116
Average number of reads per clonotype: 35.18
Reads used in clonotypes, percent of total: 74448 (87.76%)
Reads used in clonotypes before clustering, percent of total: 76478 (90.15%)
Number of reads used as a core, percent of used: 75378 (98.56%)
Mapped low quality reads, percent of used: 1100 (1.44%)
Reads clustered in PCR error correction, percent of used: 2030 (2.65%)
Reads pre-clustered due to the similar VJC-lists, percent of used: 33 (0.04%)
Reads dropped due to the lack of a clone sequence, percent of total: 4236 (4.99%)
Reads dropped due to low quality, percent of total: 116 (0.14%)
Reads dropped due to failed mapping, percent of total: 2590 (3.05%)
Reads dropped with low quality clones, percent of total: 1 (0%)
Clonotypes eliminated by PCR error correction: 832
Clonotypes dropped as low quality: 1
Clonotypes pre-clustered due to the similar VJC-lists: 27
TRB chains: 2116 (100%)
软件内置了免疫组库数据库,所以会根据提供的物种名字,自己去比对BCR的IGH,IGK,IGL这3类,以及TCR的TRA,TRB,TRD,TRG,它们各自都有V,D(可选),J,C基因。因为我们这个测试数据是TRB的,所以只有TRB的结果出来,如下:
2117 ERR3445170.clonotypes.ALL.txt
1 ERR3445170.clonotypes.IGH.txt
1 ERR3445170.clonotypes.IGK.txt
1 ERR3445170.clonotypes.IGL.txt
1 ERR3445170.clonotypes.TRA.txt
2117 ERR3445170.clonotypes.TRB.txt
1 ERR3445170.clonotypes.TRD.txt
1 ERR3445170.clonotypes.TRG.txt
理解MiXCR软件的输出结果
如下:
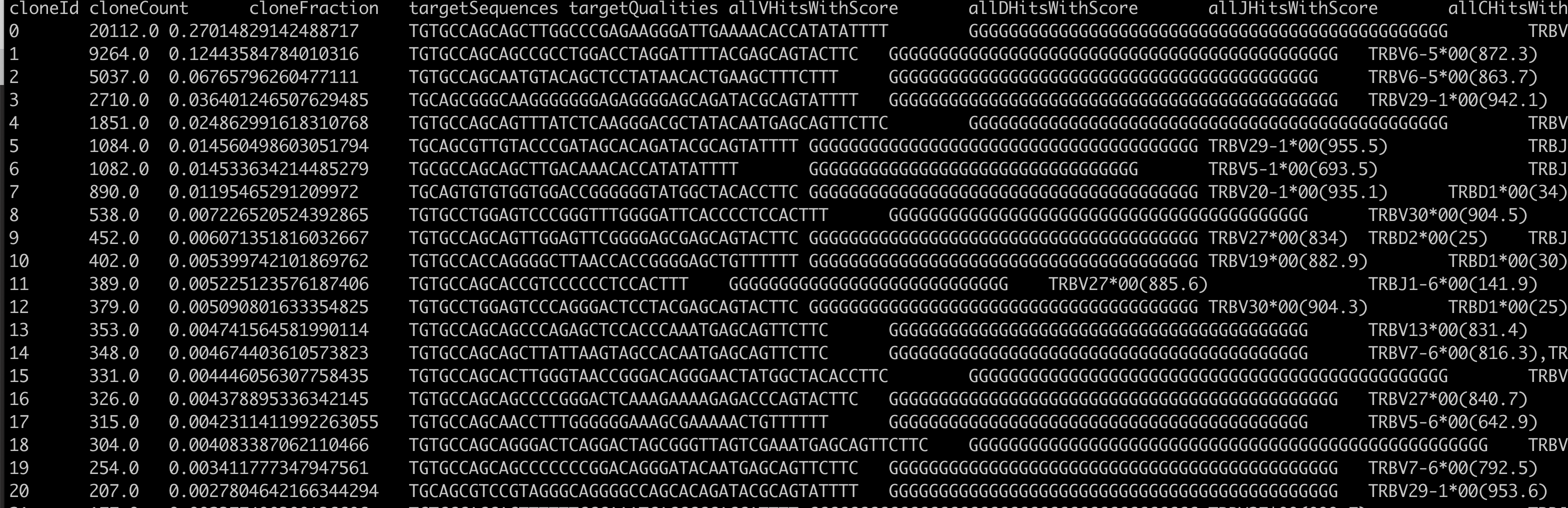
输出的列比较多:
1 cloneId
2 cloneCount
3 cloneFraction
4 targetSequences
5 targetQualities
6 allVHitsWithScore
7 allDHitsWithScore
8 allJHitsWithScore
9 allCHitsWithScore
10 allVAlignments
11 allDAlignments
12 allJAlignments
13 allCAlignments
14 nSeqFR1
15 minQualFR1
16 nSeqCDR1
17 minQualCDR1
18 nSeqFR2
19 minQualFR2
20 nSeqCDR2
21 minQualCDR2
22 nSeqFR3
23 minQualFR3
24 nSeqCDR3
25 minQualCDR3
26 nSeqFR4
27 minQualFR4
28 aaSeqFR1
29 aaSeqCDR1
30 aaSeqFR2
31 aaSeqCDR2
32 aaSeqFR3
33 aaSeqCDR3
34 aaSeqFR4
35 refPoints
其实比较容易理解,分成V,D(可选),J基因的比对情况,分成FR1-4以及CDR1-3的序列情况详情。实际上它可以把这拆分成为多个文件,这样方便浏览。
第一部分是免疫组库核心序列
因为V,D,J,C基因具有一定程度的相似性,所以并不需要全部的几百个碱基序列才能确定是V,D,J,C基因的哪些组成。
cloneId cloneCount cloneFraction targetSequences
0 20112.0 0.27014829142488717 TGTGCCAGCAGCTTGGCCCGAGAAGGGATTGAAAACACCATATATTTT
1 9264.0 0.12443584784010316 TGTGCCAGCAGCCGCCTGGACCTAGGATTTTACGAGCAGTACTTC
2 5037.0 0.06765796260477111 TGTGCCAGCAATGTACAGCTCCTATAACACTGAAGCTTTCTTT
3 2710.0 0.036401246507629485 TGCAGCGGGCAAGGGGGGGAGAGGGGAGCAGATACGCAGTATTTT
4 1851.0 0.024862991618310768 TGTGCCAGCAGTTTATCTCAAGGGACGCTATACAATGAGCAGTTCTTC
5 1084.0 0.014560498603051794 TGCAGCGTTGTACCCGATAGCACAGATACGCAGTATTTT
6 1082.0 0.014533634214485279 TGCGCCAGCAGCTTGACAAACACCATATATTTT
7 890.0 0.01195465291209972 TGCAGTGTGTGGTGGACCGGGGGGTATGGCTACACCTTC
8 538.0 0.007226520524392865 TGTGCCTGGAGTCCCGGGTTTGGGGATTCACCCCTCCACTTT
命令是:cut -f 1-4 ERR3445170.clonotypes.TRB.txt|head
第二部分是V,D,J,C基因详情
主要是方便统计该样本的V,D,J,C基因占比
allVHitsWithScore allJHitsWithScore allVAlignments allJAlignments
TRBV5-5*00(886.1) TRBJ1-3*00(197) 549|567|585|0|18||90.0 26|42|70|32|48||80.0
TRBV6-5*00(872.3) TRBJ2-7*00(175.7) 519|530|556|0|11||55.0 24|39|67|30|45||75.0
TRBV6-5*00(863.7) TRBJ1-1*00(217.9) 519|529|556|0|10||50.0 22|40|68|25|43||90.0
TRBV29-1*00(942.1) TRBJ2-3*00(204.2) 700|707|734|0|7||35.0 24|41|69|28|45||85.0
TRBV27*00(923.6) TRBJ2-1*00(200.4) 556|573|593|0|17||85.0 24|42|70|30|48||90.0
TRBV29-1*00(955.5) TRBJ2-3*00(230.1) 700|710|734|0|10||50.0 19|41|69|17|39||110.0
TRBV5-1*00(693.5) TRBJ1-3*00(195.9) 554|569|590|0|15||75.0 26|42|70|17|33||80.0
TRBV20-1*00(935.1) TRBJ1-2*00(208.9) 759|766|793|0|7||35.0 25|40|68|24|39||75.0
TRBV30*00(904.5) TRBJ1-6*00(160.6) 787|799|821|0|12||60.0 28|45|73|25|42||85.0
命令是:cut -f 6-13 ERR3445170.clonotypes.TRB.txt|head|cut -f 1,3,5,7
第3,4部分大家其实仅仅是关系CDR3区域
其中第3部分是CDR3区域的核苷酸序列,第4部分是CDR3区域的氨基酸序列。
nSeqCDR3 aaSeqCDR3
TGTGCCAGCAGCTTGGCCCGAGAAGGGATTGAAAACACCATATATTTT CASSLAREGIENTIYF
TGTGCCAGCAGCCGCCTGGACCTAGGATTTTACGAGCAGTACTTC CASSRLDLGFYEQYF
TGTGCCAGCAATGTACAGCTCCTATAACACTGAAGCTTTCTTT CASNVQL_YNTEAFF
TGCAGCGGGCAAGGGGGGGAGAGGGGAGCAGATACGCAGTATTTT CSGQGGERGADTQYF
TGTGCCAGCAGTTTATCTCAAGGGACGCTATACAATGAGCAGTTCTTC CASSLSQGTLYNEQFF
TGCAGCGTTGTACCCGATAGCACAGATACGCAGTATTTT CSVVPDSTDTQYF
TGCGCCAGCAGCTTGACAAACACCATATATTTT CASSLTNTIYF
TGCAGTGTGTGGTGGACCGGGGGGTATGGCTACACCTTC CSVWWTGGYGYTF
TGTGCCTGGAGTCCCGGGTTTGGGGATTCACCCCTCCACTTT CAWSPGFGDSPLHF
命令是:cut -f 24,33 ERR3445170.clonotypes.TRB.txt|head
也可以自己构建数据库
因为这个软件是内置了免疫组库数据库,所以会根据提供的物种名字,自己去比对BCR的IGH,IGK,IGL这3类,以及TCR的TRA,TRB,TRD,TRG,它们各自都有V,D(可选),J,C基因。
可能会出现版本更新不及时,或者你研究的物种,并不在软件考虑范围内的情况,那么你可能是需要自行构建哦。