我们前面教程:450K芯片上面的甲基化探针到底需要进行哪些过滤 已经强调过了甲基化芯片数据分析的一些注意事项,以及标准代码,共享了大量的学习资料。也分享了一篇甲基化芯片文献的标准图表,现在需要进行数据挖掘了。
我这里先列出学徒作业,大家需求下载头颈癌里面的口腔癌的甲基化芯片信号值矩阵,然后挑选有N-T配对的32个病人的数据进行差异分析,就走我们介绍的champ流程即可!
理论上你掌握了这个分析策略,换任何一个癌症,你都是可以进行同样的代码挖掘流程哦。
文章挑选32 OSCC patients
The training set including 313 OSCC cases were downloaded from the TCGA data portal accessed on March, 2016. Tumor sites of oral cavity, oral tongue, buccal mucosa, lip, alveolar ridge, hard palate, and floor of mouth were included. Patients were diagnosed during 1992– 2013, and those with missing follow-up information were excluded. Of them, 32 OSCC patients had both tumor and adjacent non-tumor tissue samples, which was used as the discovery set to identify differential methylation CpG sites.
我通常是去UCSC的xena浏览器下载指定数据,我们这个作业里面是下载头颈癌里面的口腔癌的甲基化芯片信号值矩阵,然后挑选有N-T配对的32个病人的数据进行差异分析。
文章路线图
就是下载头颈癌里面的口腔癌的甲基化芯片信号值矩阵,然后挑选有N-T配对的32个病人的数据进行差异分析,走champ流程即可!
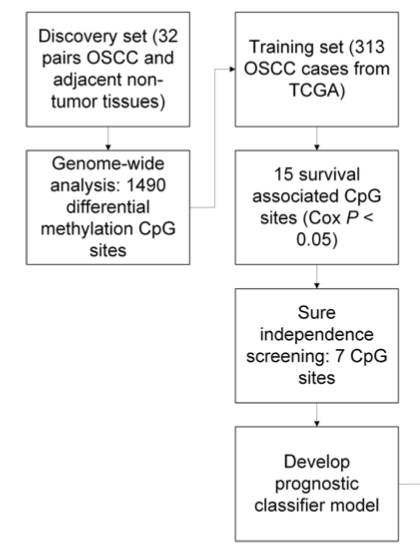
对你来说,难点可能是如何挑选到那32个病人。
TCGA的甲基化芯片信号值矩阵下载
我一般是推荐大家在 https://xenabrowser.net/datapages/ 癌症,选择头颈癌,进入下载Methylation450k (n=580) TCGA Hub
- TCGA head & neck squamous cell carcinoma (HNSC) DNA methylation data. DNA methylation profile was measured experimentally using the Illumina Infinium HumanMethylation450 platform. Beta values were derived at the Johns Hopkins University and University of Southern California TCGA genome characterization center. DNA methylation values, described as beta values, are recorded for each array probe in each sample via BeadStudio software.
- DNA methylation beta values are continuous variables between 0 and 1, representing the ratio of the intensity of the methylated bead type to the combined locus intensity.
- Thus higher beta values represent higher level of DNA methylation, i.e. hypermethylation and lower beta values represent lower level of DNA methylation, i.e. hypomethylation.
- We observed a bimodal distribution of the beta values from both methylation27 and methylation450 platforms, with two peaks around 0.1 and 0.9 and a relatively flat valley around 0.2-0.8. The bimodal distribution is far more pronounced and balanced in methylation450 than methylation27 platform. In the methylation27 platform, the lower beta peak is much stronger than the higher beta peak, while the two peaks are of similar height in the methylation450 platform. Microarray probes are mapped onto the human genome coordinates using xena probeMap derived from GEO GPL13534 record. Here is a reference to Illumina Infinium BeadChip DNA methylation platform beta value.
关于甲基化信号值矩阵,页面介绍的非常清楚了。你也可以看前面生信技能树的介绍甲基化芯片的背景知识,主要是理解什么是DNA甲基化,为什么要检测它,以及芯片和测序两个方向的DNA甲基化检测技术。具体介绍在:甲基化的一些基础知识,也了解了甲基化芯片的一般分析流程 。差异分析后的火山图
同样的,差异走champ流程哈:
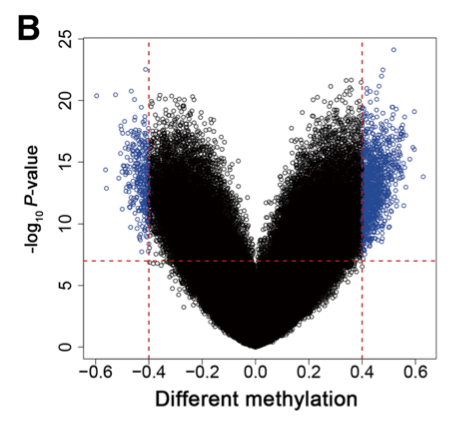
b Volcano plot comparing CpG methylation for OSCC tumor and non-tumor tissues. A total of 1490 CpG sites had an absolute value of differential methylation of > 0.4 and a paired t test P value of < 1 × 10−7 (blue dots).
注意这个阈值比较严格
生存分析对差异甲基化探针进行过滤
就是取交集啦,进一步缩小范围
差异分析后的热图
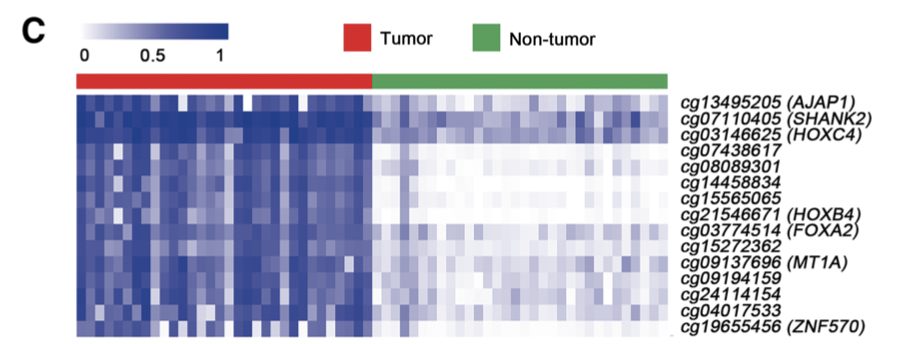
最后剩下15个甲基化探针,可以首先热图可视化N-T的差异化情况,然后生存分析看看它们的生存效果。
c Heatmap showing methylation of 15个甲基化探针 in tumor tissues and adjacent non-tumor tissues.
其它学徒作业
学徒不仅仅是在我这里学生物信息学相关知识,更重要的是能够筛选极少部分同样热爱知识整理和分享的小伙伴,所以我继续安排了一些学徒作业题:
- 学徒考核-计算wes数据的全部外显子的平均测序深度
- 为什么癌症病人据肿瘤单细胞水平的异质性但是细胞系没
- RNAseq数据,下载GEO中的FPKM文件后该怎么下游分析
- GSE83521/GSE89143数据集-需去除批次效应
- GSVA或者GSEA各种算法都是可以自定义基因集的
- limma和edgeR对RNA-seq表达矩阵差异分析的区别
- 肿瘤外显子视频课程小作业
- 为什么不用TCGA数据库来看感兴趣基因的生存情况
如果看到这些习题的你也感兴趣加入我们的在线学徒列表,欢迎随机挑选一个作业尝试完成后,邮件汇报给我你探索的过程!我的邮箱 jmzeng1314@163.com这个甲基化数据分析不免费了
因为分析起来的确很耗费计算机资源了
文末友情宣传
强烈建议你推荐给身边的博士后以及年轻生物学PI,多一点数据认知,让他们的科研上一个台阶:
- 全国巡讲全球听(买一得五) ,你的生物信息学入门课
- 生信技能树的2019年终总结 ,你的生物信息学成长宝藏
- 2020学习主旋律,B站74小时免费教学视频为你领路