本质上是使用发表在 Nat Methods. 2015 May;的CIBERSORT算法,对TCGA数据库的RNA-seq数据,计算并构建了一个数据库网页工具: The Cancer Immunome Atlas (https://tcia.at/)
这个网页工具发表在 Cell Rep. 2017 Jan 3;1 标题是:Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade.
其workflow如下:
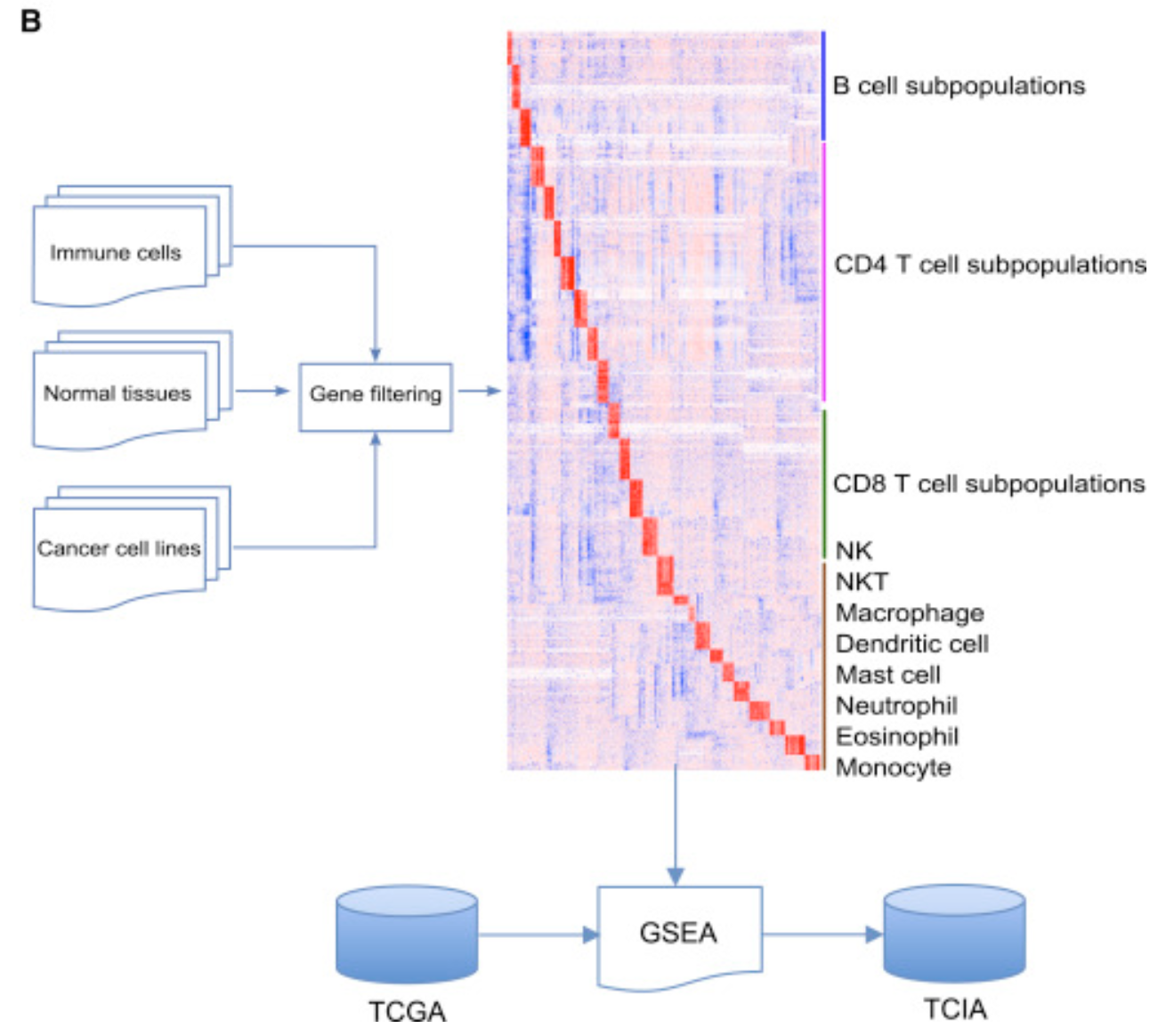
其中这28 subpopulations of TILs可以分成2大类:
- adaptive immunity: activated T cells, central memory (Tcm), effector memory (Tem) CD4+ and CD8+ T cells, gamma delta T (Tγδ) cells, T helper 1 (Th1) cells, Th2 cells, Th17 cells, regulatory T cells (Treg), follicular helper T cells (Tfh), activated, immature, and memory B cells
- innate immunity, such as macrophages, monocytes, mast cells, eosinophils, neutrophils, activated, plasmacytoid, and immature dendritic cells (DCs), NK cells, natural killer T (NKT) cells, and MDSCs.
这28个细胞数据来源: Download spreadsheet (22KB) Table S5. List of Microarray Data from 37 Studies for Identifying Immune-Related Genes.
全部的基因列表:Download spreadsheet (32KB) Table S6. List of Pan-cancer Immune Metagenes.