这篇文章最重要的观点是甲基化用来给GBM分组,分成了6组,至于突变什么的,随便讲了讲,反正数据也不给下载。
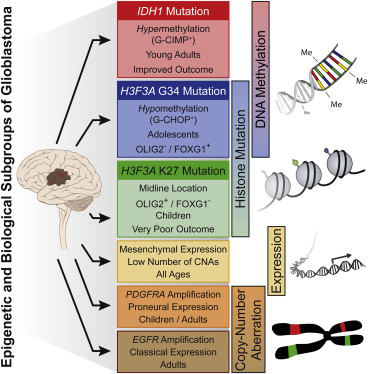
We identified six epigenetic and biological GBM subgroups displaying distinct global DNA methylation patterns, which harbor unique hotspot mutations, DNA copy-number alterations, and transcriptomic patterns.
分组如下:
主要是做甲基化(GSE36278)
We investigated a cohort of GBMs from children (n = 59) and adult patients (n = 77) for genome-wide DNA methylation patterns using the Illumina 450k methylation array.
Consensus clustering using the 8,000 most variant probes across the data set robustly identified six distinct DNA methylation clusters.
Based on correlations with mutational status, DNA copy-number aberrations, and gene expression signatures, as outlined below, we have labeled these subgroups “IDH,” “K27,” “G34,” “RTK I (PDGFRA),” “Mesenchymal,” and “RTK II (Classic).”
还根据肿瘤部位,一下临床marker,还有生存情况来论证它分组的正确性。
样本量是:
Primary tumor samples for methylation (n = 136; Table S1), mutation (n = 460; Table S2), and gene expression (n = 69) analysis and all clinical data were collected at the DKFZ (Heidelberg, Germany) and at McGill University (Montreal, Canada).
测序用的是杂交捕获PCR纯化测序方法。
表达量增加了46个样本!
数据地址是;
The complete CpG methylation values are accessible through GEO Series accession number GSE36278.
The complete gene expression values are accessible through GEO Series accession numbers GSE36245 and, as part of a previously reported series, GSE34824 (Schwartzentruber et al., 2012).