一般做完一个CHIP-seq测序,如果实验设计没有问题,测序质量也OK的话,很容易了根据序列call到符合要求的peaks,或者可以去很多文章或者roadmap里面下载到非常多有意义的peaks文件, 一般是BED格式文件,这是就需要对这些peaks进行各种各样的注释以及可视化了,还有根据peaks相关的基因可以做各种各样的下游分析,包括各种pathway数据库的富集,MsigDB数据库注释,gene ontology的注释等等,此时不得不强烈推荐一款网页版工具,是斯坦福大学的学者开发的GREAT。
此工具的出现主要是为了解决基因组上面的非编码区域注释缺乏的问题,而我们CHIP-seq实验得到的peaks结果通常就是在非编码区域
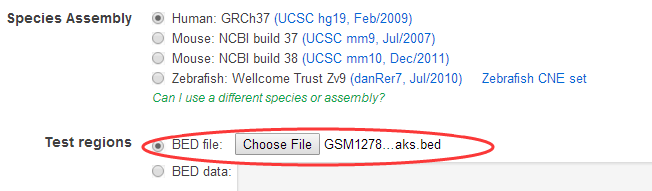
该工具每次只能上传一个文件,就是我们call出来的peaks记录文件,支持bed格式的:
一般很快就可以出结果啦!
首先会有三个图,都是很常见的,大家随便看看咯
Number of associated genes per region
Binned by orientation and distance to TSS
Binned by absolute distance to TSS
然后就是pathway和GO注释啦
这个网站提供的pathway非常之多,还是蛮全面的,包括KEGG,biocarta,reactome,msigdb等等还有一些signature和gene families,相当于一站式完成了大部分下游分析
GO Molecular Function (no terms)
GO Biological Process (no terms)
GO Cellular Component (no terms)
The test set of 5,225 genomic regions picked 2,992 (17%) of all 18,041 genes.
GO Molecular Function has 3,688 terms covering 15,090 (84%) of all 18,041 genes, and 189,388 term - gene associations.
3,688 ontology terms (100%) were tested using an annotation count range of [1, Inf].
The test set of 5,225 genomic regions picked 2,992 (17%) of all 18,041 genes.
GO Biological Process has 10,440 terms covering 15,441 (86%) of all 18,041 genes, and 950,065 term - gene associations.
10,440 ontology terms (100%) were tested using an annotation count range of [1, Inf].
The test set of 5,225 genomic regions picked 2,992 (17%) of all 18,041 genes.
GO Biological Process has 10,440 terms covering 15,441 (86%) of all 18,041 genes, and 950,065 term - gene associations.
GO Biological Process has 10,440 terms covering 15,441 (86%) of all 18,041 genes, and 950,065 term - gene associations.
10,440 ontology terms (100%) were tested using an annotation count range of [1, Inf].
Mouse Phenotype (no terms)
Human Phenotype (no terms)
Disease Ontology (no terms)
MSigDB Cancer Neighborhood (no terms)
Placenta Disorders (no terms)
PANTHER Pathway (no terms)
BioCyc Pathway (no terms)
MSigDB Pathway (no terms)
MGI Expression: Detected (no terms)
MSigDB Perturbation (no terms)
MSigDB Predicted Promoter Motifs (no terms)
MSigDB miRNA Motifs (no terms)
InterPro (no terms)
InterPro (no terms)
HGNC Gene Families (no terms)
MSigDB Oncogenic Signatures (no terms)
MSigDB Immunologic Signatures (no terms)
The test set of 5,225 genomic regions picked 2,992 (17%) of all 18,041 genes.
MSigDB Immunologic Signatures has 1,910 terms covering 16,609 (92%) of all 18,041 genes, and 363,333 term - gene associations.
1,910 ontology terms (100%) were tested using an annotation count range of [1, Inf].