2017的一个文章:《Meta-signature of human endometrial receptivity: a meta- analysis and validation study of transcriptomic biomarkers》 做了很多独立的公共数据集里面的表达量矩阵分组后的差异分析,然后使用rra整合算法拿到了可靠的上下调基因,但是隐藏在下面的图里面:
我的提问非常简单:
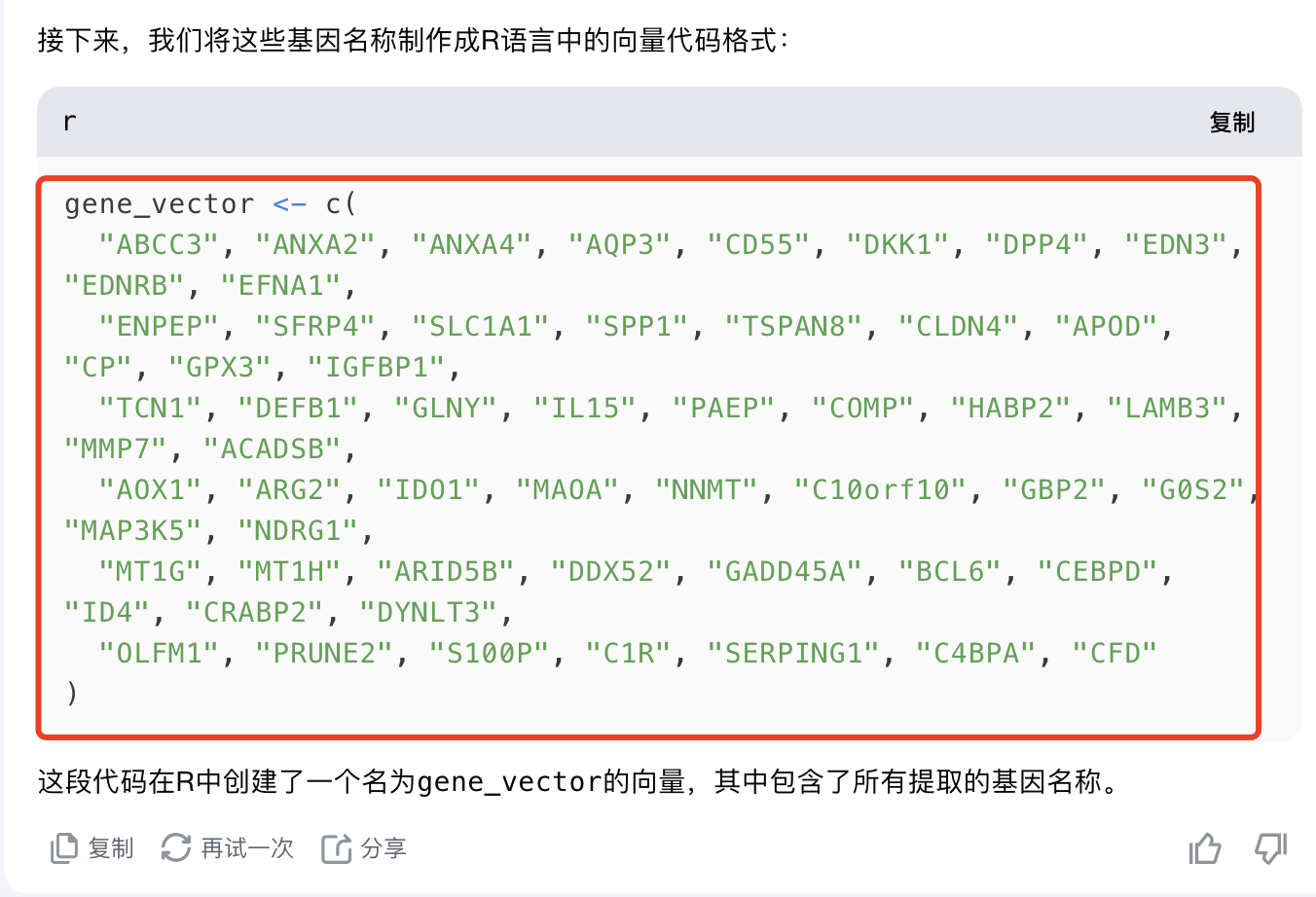
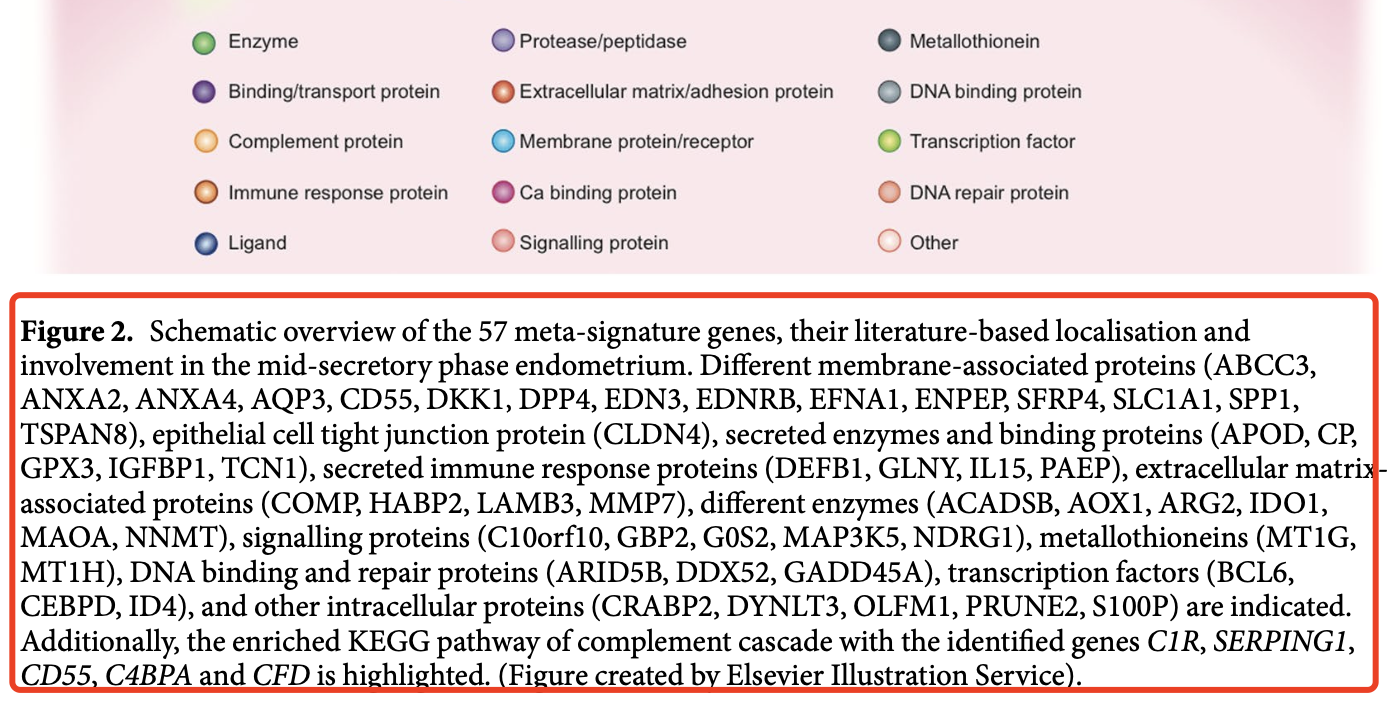
帮我把里面的基因名字提取出来,然后制作成为了r里面的向量的代码格式: Different membrane-associated proteins (ABCC3, ANXA2, ANXA4, AQP3, CD55, DKK1, DPP4, EDN3, EDNRB, EFNA1, ENPEP, SFRP4, SLC1A1, SPP1, TSPAN8), epithelial cell tight junction protein (CLDN4), secreted enzymes and binding proteins (APOD, CP, GPX3, IGFBP1, TCN1), secreted immune response proteins (DEFB1, GLNY, IL15, PAEP), extracellular matrix- associated proteins (COMP, HABP2, LAMB3, MMP7), different enzymes (ACADSB, AOX1, ARG2, IDO1, MAOA, NNMT), signalling proteins (C10orf10, GBP2, G0S2, MAP3K5, NDRG1), metallothioneins (MT1G, MT1H), DNA binding and repair proteins (ARID5B, DDX52, GADD45A), transcription factors (BCL6, CEBPD, ID4), and other intracellular proteins (CRABP2, DYNLT3, OLFM1, PRUNE2, S100P) are indicated. Additionally, the enriched KEGG pathway of complement cascade with the identified genes C1R, SERPING1, CD55, C4BPA and CFD is highlighted.
如下所示的对话:

而且很容易就写出来了代码: