交流群有小伙伴希望我们《生信菜鸟团》的单细胞周更专辑作者发现这个新鲜出炉的肝癌单细胞文章,标题是:《CD36+ cancer-associated fibroblasts provide immunosuppressive microenvironment for hepatocellular carcinoma via secretion of macrophage migration inhibitory factor》,简单的看了看里面的图表,就是降维聚类分群后,针对肝癌里面的成纤维细胞亚群进行细分和讲解,数据集链接是;
可以看到是 7 HBV-related HCC tissues and 4 adjacent liver tissues 的总共11个单细胞转录组样品,样品具体信息如下所示:
GSM6127499 Human hepatocellular carcinoma tissues [2a-0706]
GSM6127500 Human hepatocellular carcinoma tissues [1a-0618]
GSM6127501 Human hepatocellular carcinoma tissues [1a-0629]
GSM6127502 Human hepatocellular carcinoma tissues [1a-0706]
GSM6127503 Human hepatocellular carcinoma tissues [1a-0707]
GSM6127504 Human hepatocellular carcinoma tissues [1a-0727]
GSM6127505 Human hepatocellular carcinoma tissues [1a-0831]
GSM6127506 Human adjacent liver tissues [1b-0618]
GSM6127507 Human adjacent liver tissues [2b-0618]
GSM6127508 Human adjacent liver tissues [X-1b]
GSM6127509 Human adjacent liver tissues [B-2b]
但是它给出来矩阵的时候是混合的,其实也很简单就可以读取的,我们《生信菜鸟团》的单细胞周更专辑作者分享过好几次了基础文件读取技巧啦,详见: 读取不同格式的单细胞转录组数据及遇到问题的解决办法
GSE202642_barcodes.tsv.gz 569.6 Kb
GSE202642_features.tsv.gz 325.6 Kb
GSE202642_matrix.mtx.gz 666.5 Mb
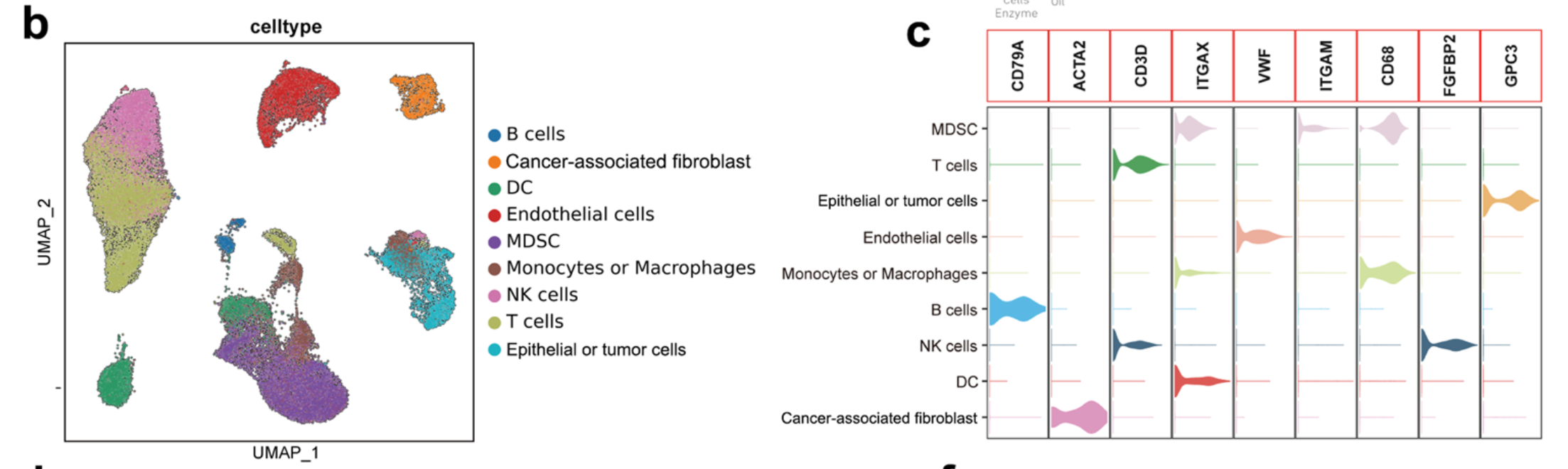
可以看到作者的第一层次降维聚类分群是9大亚群 :
- epithelial and tumor cells (5059 cells, 5.6%, marked with EPCAM, ALDH1A1 and ALB)
- B cells (1332 cells, 1.5%, marked with MS4A1 and CD79A)
- T cells (24895 cells, 27.5%, marked with CD3D and CD3E)
- natural killer (NK) cells (13868 cells, 15.3%, marked with FGFBP2 and FCG3RA)
- myeloid-derived suppressor cells (MDSCs) (11087 cells, 12.2%, marked with ITGAM and CD33)
- monocytes or macrophages (9978 cells, 11.0%, marked with CD68, CD163, and CD14)
- dendritic cells (7784 cells, 8.6%, marked with ITGAX)
- fibroblasts (2495 cells, 2.8%, marked with ACTA2 and COL1A2)
- endothelial cells (14074 cells, 15.5%, marked with PECAM1 and vWF;
同样的是肝癌,并没有出现NK细胞占比超50%的情况,但是前面我们分享的 SingleR说是NK细胞你就相信了吗,数据集GSE162616里面的3个样品的单细胞数据,就Cells in clusters 0, 1, 2, 3, 7, 9, 10, 12, and 14 were annotated as NK cells based on SingleR ,也就是说占比超过了50%,要么是数据集GSE162616有问题,要么是这个数据集GSE162616的数据挖掘文章作者处理有问题。
前面的数据集GSE202642的第一层次降维聚类分群是9大亚群可视化如下所示:
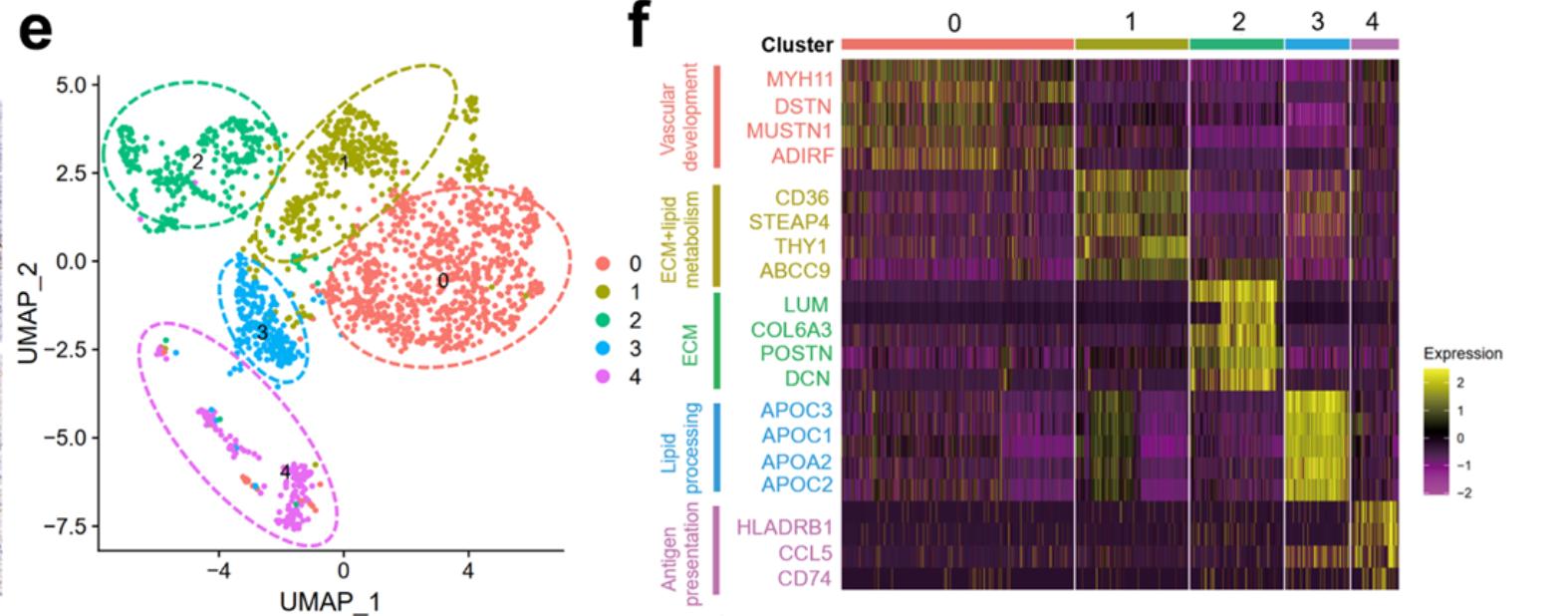
文章的分析很简单,,就是降维聚类分群后,针对肝癌里面的成纤维细胞亚群进行细分和讲解:
可以看到作者把肝癌里面的成纤维细胞亚群分成了5个并且进行了功能性讲解,也就是常规的go或者kegg数据库注释啦, 就有了5个亚群的名字(vCAFs, mCAFs, lpmCAFs (CD36+ CAFs), lpCAFs and apCAFs,),如下所示:
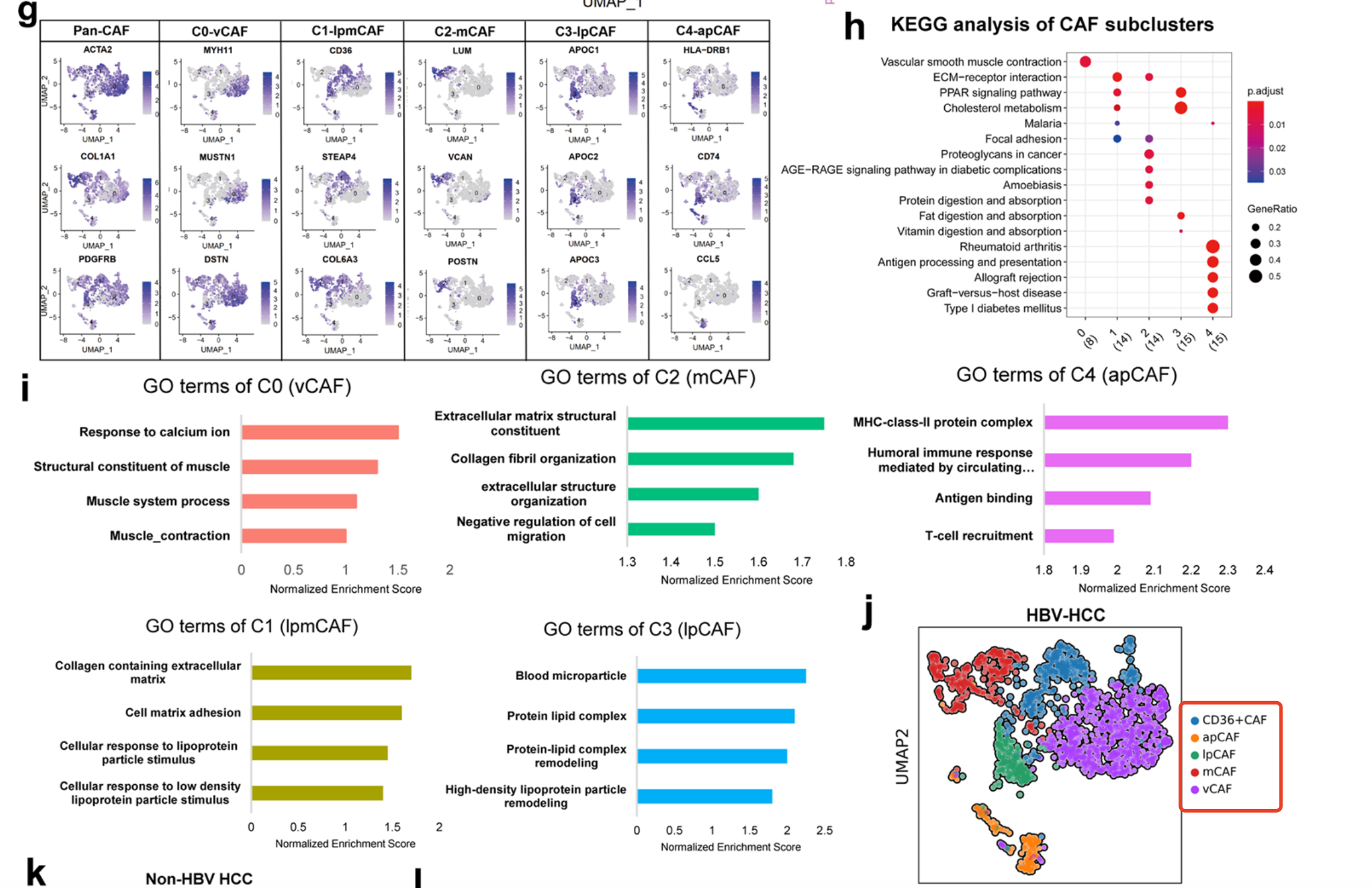
而且使用了小鼠单细胞数据集去验证了:
- To further discern fibroblast heterogeneity, we then sorted 1746 CAFs in our scRNA-seq analyses derived from 7 murine HCC tumors and clustered the CAFs into 7 subpopulations
当然了,因为物种不一样,所以肝癌病人的单细胞跟小鼠是有差异,但是绝大部分情况还是类似的:
- we found that all 5 CAF subtypes identified in human HCC tumors were also represented in mouse HCC tissues
- the 5 mouse CAF subsets possessed populations and gene signatures similar to those of human CAFs, including vCAFs, mCAFs, CD36+CAFs, lpCAFs and apCAFs
早期就有很多其它癌症也有成纤维单细胞亚群解析
甚至有泛癌层面,比如2022年底的:《Pan-cancer single-cell analysis reveals the heterogeneity and plasticity of cancer-associated fibroblasts in the tumor microenvironment》,就是对 10 种实体癌类型的 226 个样本进行泛癌分析,以单细胞分辨率分析 TME,说明异质癌症相关成纤维细胞 (CAF) 的共性/可塑性:
- 148 primary tumor, 53 adjacent, and 25 normal samples from 164 donors enrolled in 12 studies
- 855,271 cells from 226 samples were included and integrated based on a batch effect correction algorithm
- 34 TME-related clusters (c1-c34, 569,759 cells),
- fibroblasts (c1-c8, DCN, COL1A1),
- lymphocytes (c9-c17, CD3D, CD3E),
- myeloid cells (c18-c24, CD68, CD14),
- endothelial cells (c25-c28, VWF, PECAM1),
- plasma cells (c29-c34, IGHG1, JCHAIN)
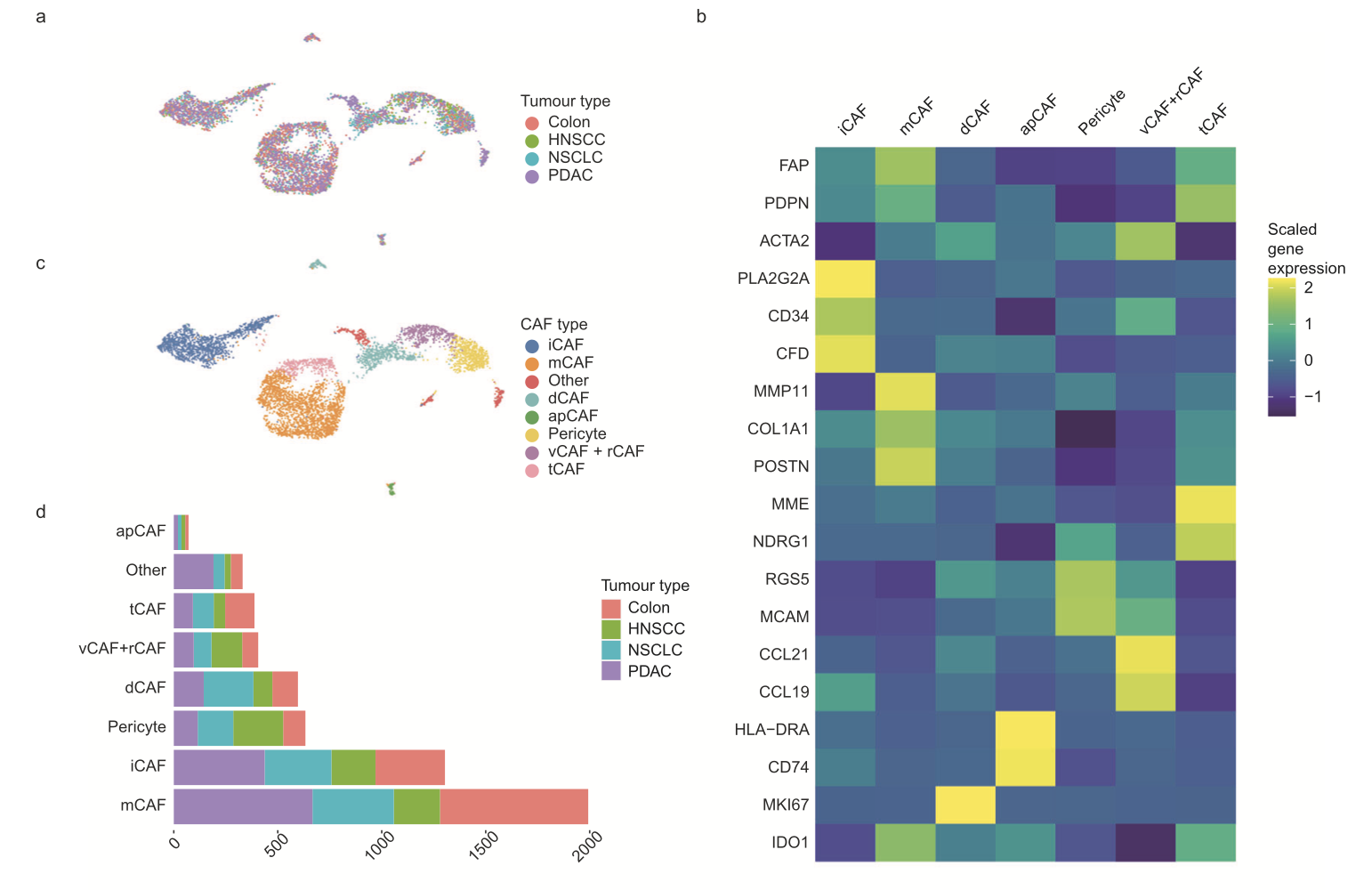
也有具体的癌症层面的单细胞数据挖掘文章,比如同样是2023发表的数据挖掘文章:《Cancer-associated fibroblast classification in single-cell and spatial proteomics data》,分析了已经发表的来自14名乳腺癌患者肿瘤组织的约11万细胞里面的16,704个基质细胞,分群12数量亚群后合并为10个生物学意义亚群,分别是(9 CAF types and one cluster of pericytes) ,最后的单细胞亚群的各自的基因如下所示:
- RGS5 for pericytes
- MCAM for vCAFs
- MME (together with NDRG1 and ENO1) for tCAFs
- IDO1 for ifnCAFs
- HSPH1 for hsp_tCAFs
- MMP11 (together with COL1A1 and POSTN) for mCAFs
- PLA2G2A, CDF and CD34 for iCAFs
- MKI67 for dCAFs
- CCL21 (and CCL19) for rCAFs
- CD74 and HLA-DR for apCAFs
- PDPN and FAP for the general category of activated CAFs
而且也是在多个癌症里面的检验了,可视化如下所示:
所以最后有成纤维单细胞亚群规范命名标准吗
可以看到,每个癌症的单细胞数据集分析结果,多个癌症整合的结果,不同的物种,其实最后得到的单细胞亚群非常难以对比。但是如果是我们说b细胞和t细胞,就没有癌症没有物种的考虑,当然了如果是具体到b细胞里面的细分亚群,或者t细胞里面的细分亚群,也会面临同样的问题就是跨越数据集就很难去比较了。