文章:《Preparation of mouse pancreatic tumor for single-cell RNA sequencing and analysis of the data》,链接:https://www.sciencedirect.com/science/article/pii/S266616672100695X
分享了一个Bioinformatics analysis of single-cell RNA sequencing,其封装了数据分析流程成为了一个 reproduc- ible Common Workflow Language (CWL) pipelines ,而且可以在 user-friendly Scientific Data Analysis Platform (SciDAP, https://scidap.com). 平台直接运行。流程示意图如下所示:
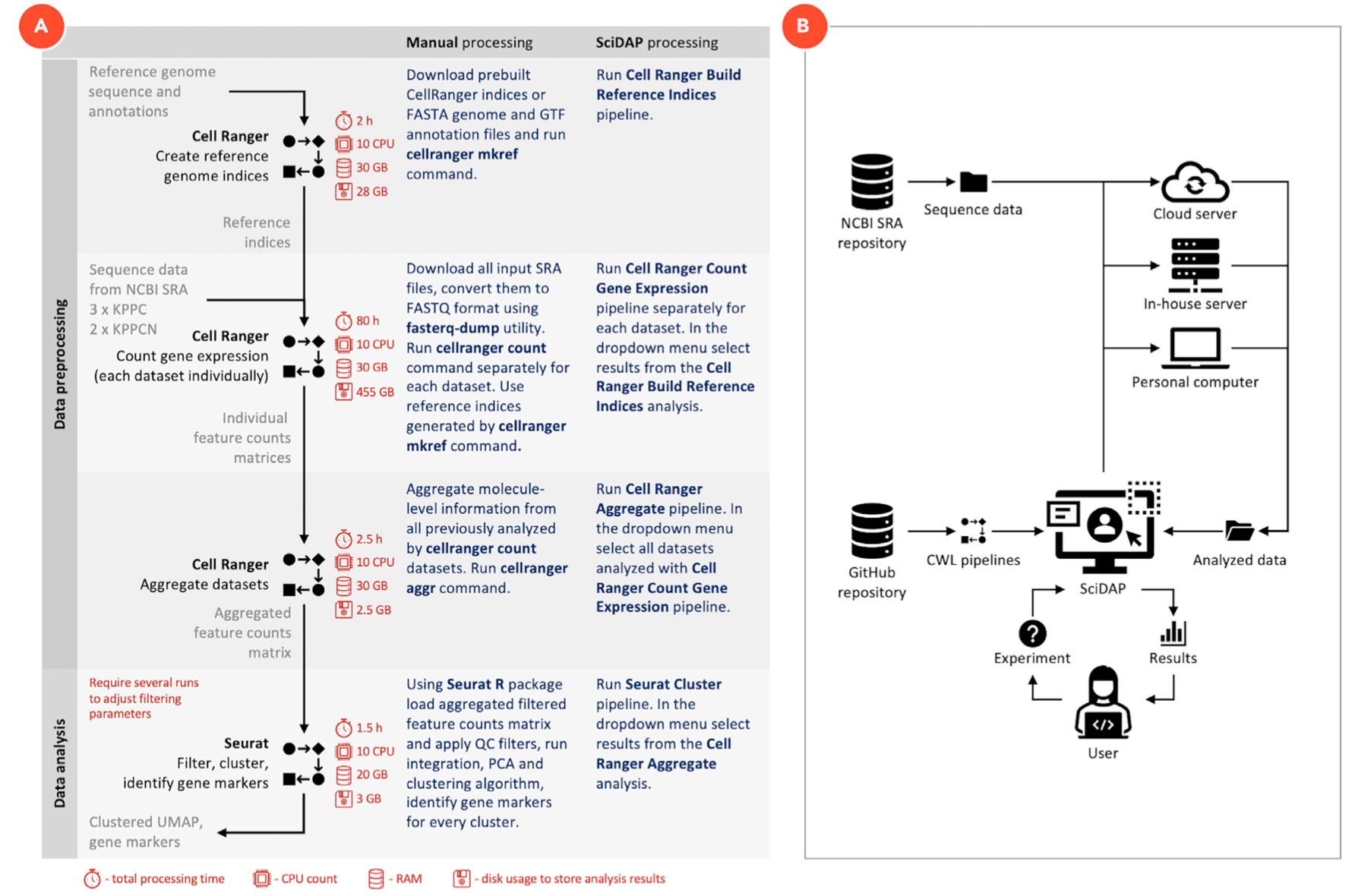
有意思的是该文献还图文并茂的介绍了如何一步步从公共数据库里面的fastq文件开始,跑完整个单细胞数据分析流程。
主要是下面的4个步骤:
- i. Cell Ranger Build Reference Indices
- ii. Cell Ranger Count Gene Expression
- iii. Cell Ranger Aggregate
- iv. Seurat Cluster
文章举例的数据是KPPC 1 SRR12450154 dataset,链接是 :https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA657051
可以看到是5个单细胞样品,物种是 小鼠 :
1.GSM4726799: KPPCN 2; Mus musculus; RNA-Seq
run: 495.7M spots, 148.7G bases, 64.6Gb downloads
Accession: SRX8944617
2.GSM4726798: KPPCN 1; Mus musculus; RNA-Seq
run: 200.3M spots, 60.1G bases, 26.1Gb downloads
Accession: SRX8944616
3.GSM4726797: KPPC 3; Mus musculus; RNA-Seq
run: 491.9M spots, 147.6G bases, 62.7Gb downloads
Accession: SRX8944615
4.GSM4726796: KPPC 2; Mus musculus; RNA-Seq
run: 488.8M spots, 146.6G bases, 63.3Gb downloads
Accession: SRX8944614
5.GSM4726795: KPPC 1; Mus musculus; RNA-Seq
run: 477.4M spots, 143.2G bases, 62.1Gb downloads
Accession: SRX8944613
Cancer Cell, 2020 Oct ,标题:《Cholesterol Pathway Inhibition Induces TGF-β Signaling to Promote Basal Differentiation in Pancreatic Cancer》,链接在:https://pubmed.ncbi.nlm.nih.gov/32976774/,数据是[GSE156210](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE156210),五个单细胞样品的表达量矩阵都是存在的:
GSM4726795 KPPC 1
GSM4726796 KPPC 2
GSM4726797 KPPC 3
GSM4726798 KPPCN 1
GSM4726799 KPPCN 2