最近课题组的文献分享交流,有一个小伙伴讲的是最近的一篇nature文章:《ecDNA hubs drive cooperative intermolecular oncogene expression》, 有点复杂,不仅仅是有多组学,还有单细胞。如下所示:
GSE159972 [ChIP]
GSE159985 [HiChIP]
GSE160148 [scRNA and scATAC-Seq]
GSE175451 [ATAC-Seq]
GSE175452 [Hi-C]
GSE184566 [RNA-Seq]
但是我看了看数据集:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE160148 ,以及其数据分析结果在文章里面的图表,居然发现它总共是16个10X单细胞样品分成两组,居然就做一个降维聚类分群并且展示了一下!
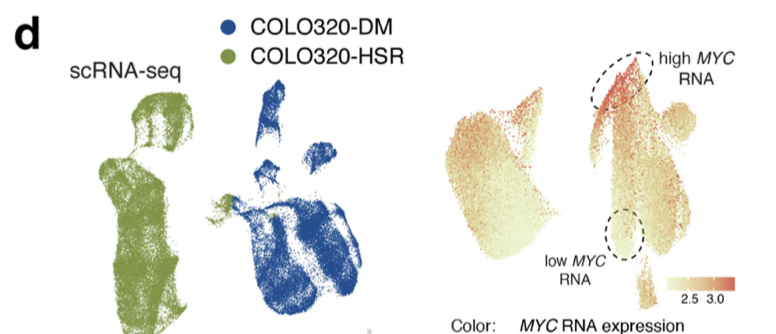
这个图首先并不是在主图里面,而是 Extended Data Fig. 5 ,其次,它仅仅是 图d的一半而已:
d, UMAP from the RNA or the ATAC–seq data (left). Log-normalized and scaled MYC RNA expression (top right) and MYC accessibility scores (bottom right) were visualized on the ATAC–seq UMAP, showing cell-level heterogeneity in MYC RNA-seq and ATAC-seq signals in ecDNA-
containing COLO320-DM.
上半部分是单细胞转录组数据的降维聚类分群,下半部分是 单细胞ATAC的结果,大家感兴趣可以去看原文!
非常容易下载其表达量矩阵文件,并且读入到R里面,安装我们的标准 代码进行降维聚类分群,跟文章进行对比!
GSE160148_scRNA_COLO320.mtx.gz 1.1 Gb (ftp)(http) MTX
GSE160148_scRNA_COLO320_barcodes.tsv.gz 523.0 Kb (ftp)(http) TSV
GSE160148_scRNA_COLO320_features.tsv.gz 200.3 Kb (ftp)(http) TSV
以前我们做了一个投票:可视化单细胞亚群的标记基因的5个方法,下面的5个基础函数相信大家都是已经烂熟于心了:
- VlnPlot(pbmc, features = c(“MS4A1”, “CD79A”))
- FeaturePlot(pbmc, features = c(“MS4A1”, “CD79A”))
- RidgePlot(pbmc, features = c(“MS4A1”, “CD79A”), ncol = 1)
- DotPlot(pbmc, features = unique(features)) + RotatedAxis()
- DoHeatmap(subset(pbmc, downsample = 100), features = features, size = 3)
其实这个文章配图的右边就是简单的 FeaturePlot 去可视化 MYC这个基因的表达量,至于配图的左边就是降维聚类分群而已!