精准医疗时代,对于癌症我们最重要的就是分而治之,理论上应该是每个人的癌症都不一样,但实际上的医疗现实不允许我们对每个癌症患者都进行事无巨细的科研探索来给他制定个性化诊疗方案。能把以前大家混完一谈的单一器官癌症区分成为不同细分癌症,就已经是科研界近几十年的成果了。
比如是乳腺癌,你可以看lumA,lumB,basal,HER2 等亚型,其中TNBC可以继续细分为3~7种亚型,当然了,现在有了单细胞转录组数据的加持,细胞亚型会越来越清晰。如果要整合多组学数据,分类也会更加复杂。
再比如是胃癌,也是有4种分子分型,具体如下:
-
①爱泼斯坦-巴尔(Epstein-Barr)病毒(EBV)阳性型肿瘤:约占胃癌的9%,表现为较高频率的PIK3CA基因突变和DNA极度超甲基化,以及JAK2、CD274(也称PD-L1)和PDCD1LG2(也称PD-L2)基因扩增。
-
②微卫星不稳定(MSI)型:约占22%,表现为重复DNA序列突变增加,包括编码靶向致癌信号蛋白的基因突变。
-
③基因稳定(GS)型:约占20%,其组织学变异弥漫且丰富,RHOA基因突变或RHO家族GTP酶活化蛋白基因融合现象多见。
-
④染色体不稳定(CIN)型:此类肿瘤占胃癌的比例近一半,表现为显著异倍体性及受体酪氨酸激酶的局部扩增。
这里,分享一个很有意思的R包:CancerSubtypes
发表这个CancerSubtypes包的文章是: (2017). “CancerSubtypes: an R/Bioconductor package for molecular cancer subtype identification, validation, and visualization.” Bioinformatics. https://doi.org/10.1093/bioinformatics/btx378.
它里面集成了6种常见的肿瘤转录组测序数据的分子分型的算法,包括:
- Consensus clustering (CC) (Monti et al., 2003)
- Consensus non-negative matrix factorization (CNMF) (Brunet et al., 2004)
- Integrative clustering (iCluster) (Shen et al., 2009)
- Similarity network fusion (SNF) (Wang et al., 2014)
- Weighted SNF (WSNF) (Xu et al., 2016)
可以看到其实都有一定年代了哦!
大家在分析TCGA数据库的时候,首先需要下载TCGA的33种癌症的全部数据,尤其是表达量矩阵和临床表型信息啦,这里我们推荐在ucsc的xena里面下载:https://xenabrowser.net/datapages/
GDC TCGA Acute Myeloid Leukemia (LAML) (15 datasets)
GDC TCGA Adrenocortical Cancer (ACC) (14 datasets)
GDC TCGA Bile Duct Cancer (CHOL) (14 datasets)
GDC TCGA Bladder Cancer (BLCA) (14 datasets)
GDC TCGA Breast Cancer (BRCA) (20 datasets)
GDC TCGA Cervical Cancer (CESC) (14 datasets)
GDC TCGA Colon Cancer (COAD) (15 datasets)
GDC TCGA Endometrioid Cancer (UCEC) (15 datasets)
GDC TCGA Esophageal Cancer (ESCA) (14 datasets)
GDC TCGA Glioblastoma (GBM) (15 datasets)
GDC TCGA Head and Neck Cancer (HNSC) (14 datasets)
GDC TCGA Kidney Chromophobe (KICH) (14 datasets)
GDC TCGA Kidney Clear Cell Carcinoma (KIRC) (15 datasets)
GDC TCGA Kidney Papillary Cell Carcinoma (KIRP) (15 datasets)
GDC TCGA Large B-cell Lymphoma (DLBC) (14 datasets)
GDC TCGA Liver Cancer (LIHC) (14 datasets)
GDC TCGA Lower Grade Glioma (LGG) (14 datasets)
GDC TCGA Lung Adenocarcinoma (LUAD) (15 datasets)
GDC TCGA Lung Squamous Cell Carcinoma (LUSC) (15 datasets)
GDC TCGA Melanoma (SKCM) (14 datasets)
GDC TCGA Mesothelioma (MESO) (14 datasets)
GDC TCGA Ocular melanomas (UVM) (14 datasets)
GDC TCGA Ovarian Cancer (OV) (15 datasets)
GDC TCGA Pancreatic Cancer (PAAD) (14 datasets)
GDC TCGA Pheochromocytoma & Paraganglioma (PCPG) (14 datasets)
GDC TCGA Prostate Cancer (PRAD) (14 datasets)
GDC TCGA Rectal Cancer (READ) (15 datasets)
GDC TCGA Sarcoma (SARC) (14 datasets)
GDC TCGA Stomach Cancer (STAD) (15 datasets)
GDC TCGA Testicular Cancer (TGCT) (14 datasets)
GDC TCGA Thymoma (THYM) (14 datasets)
GDC TCGA Thyroid Cancer (THCA) (14 datasets)
GDC TCGA Uterine Carcinosarcoma (UCS) (14 datasets)
这些癌症目前的分类仍然是太粗糙了,推荐使用 TCGAbiolinks 包获取各个癌症的细分亚群信息,代码如下:
suppressMessages(library(TCGAbiolinks))
suppressMessages(library(tidyverse))
#下载数据
phe <- as.data.frame(TCGAquery_subtype(tumor = "brca"))
table(phe$BRCA_Subtype_PAM50)
Basal Her2 LumA LumB NA Normal
192 82 562 209 2 40
也可以简单检查一下这个亚型与其它临床信息的关系:
library(gplots)
balloonplot(table(
phe$BRCA_Subtype_PAM50,
phe$pathologic_stage
))
如下所示:
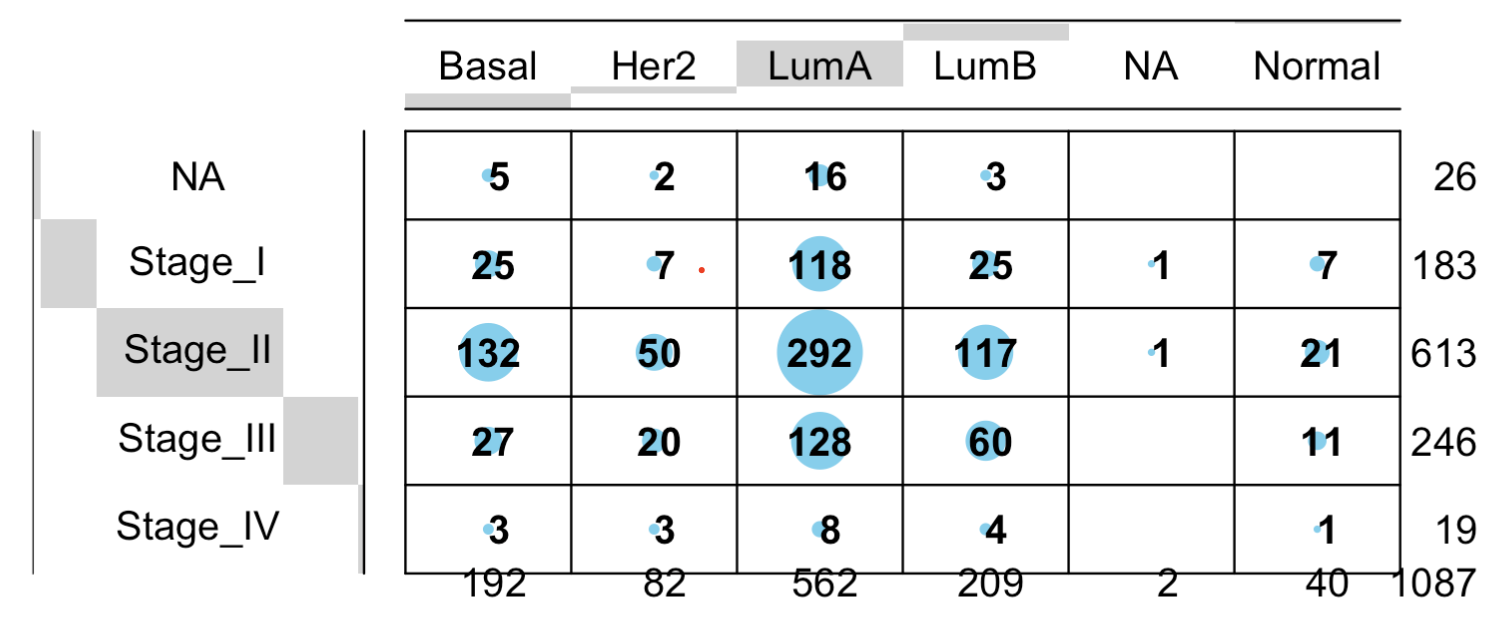
因为这个函数获取的临床信息 太多了,我们就不一一举例啦:
> colnames(phe)
[1] "patient" "Tumor.Type"
[3] "Included_in_previous_marker_papers" "vital_status"
[5] "days_to_birth" "days_to_death"
[7] "days_to_last_followup" "age_at_initial_pathologic_diagnosis"
[9] "pathologic_stage" "Tumor_Grade"
[11] "BRCA_Pathology" "BRCA_Subtype_PAM50"
[13] "MSI_status" "HPV_Status"
[15] "tobacco_smoking_history" "CNV Clusters"
[17] "Mutation Clusters" "DNA.Methylation Clusters"
[19] "mRNA Clusters" "miRNA Clusters"
[21] "lncRNA Clusters" "Protein Clusters"
[23] "PARADIGM Clusters" "Pan-Gyn Clusters"
>
可以看到R语言真的是超级方便,适合做数据挖掘啊!比如我的4个小时TCGA肿瘤数据库知识图谱视频教程,中共使用了四种算法构建模型:
- cox(可做单因素和多因素)
TCGA的cox模型构建和风险森林图 - lasso回归
用lasso回归构建生存模型+ROC曲线绘制 - 随机森林
听起来很霸气用起来并不难的随机森林 - 支持向量机
听起来很霸气用起来并不难的 支持向量机
不管用了那种算法,核心都只是几句代码而已。
其它癌症类似,只需要变化它里面的缩略词即可,感兴趣的小伙伴可以把全部的癌症都试一次。