我们的《单细胞文献100》活动激起了大家对单细胞的兴趣, 马上交流群有朋友咨询B细胞的细分亚群,希望给出参考文献,我恰好在看新冠病毒相关单细胞数据分析文章, 就给出来了这个:《Single-cell landscape of immunological responses in patients with COVID-19》
该研究的实验设计很清晰,就是 5 个正常人加上13个患者 的 血液进行单细胞转录组 :
- healthy donors (HDs) (*n* = 5),
- moderate (*n* = 7),
- severe (*n* = 4)
- convalescent (*n* = 6) samples
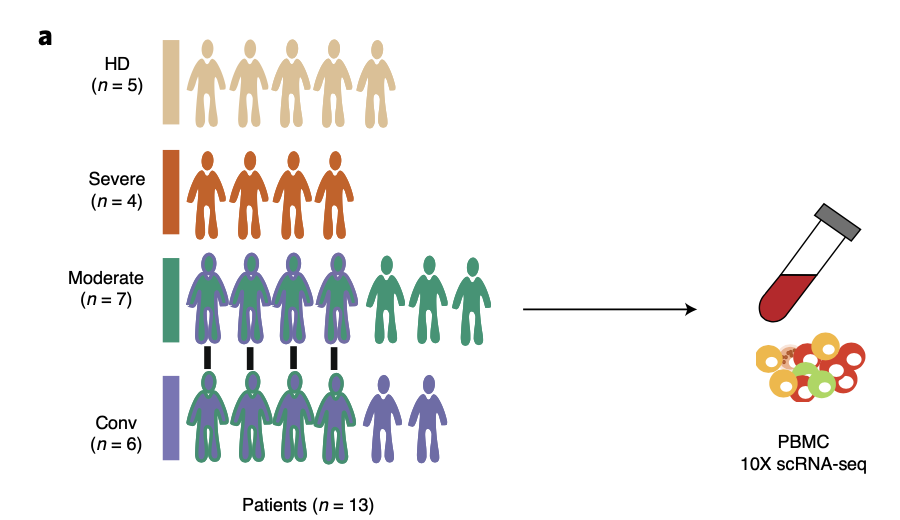
### 第一层次降维聚类分群
总共是 122,542 cells ,第一层次降维聚类分群是14个:
- naive-state T (naive T) cells (CD3+CCR7+)
- activated-state T (activated T) cells (CD3+PRF1+)
- mucosal-associated invariant T (MAIT) cells (SLC4A10+TRAV1-2+)
- γδ T cells (TRGV9+TRDV2+)
- proliferative T (pro T) cells (CD3+MKI67+)
- natural killer (NK) cells (KLRF1+)
- B cells (MS4A1+)
- plasma B cells (MZB1+)
- CD14+ monocytes (CD14+ mono; LYZ+CD14+)
- CD16+ monocytes (CD16+ mono; LYZ+FCGR3A+)
- monocyte-derived dendritic cells (mono DCs; CD1C+)
- plasmacytoid dendritic cells (pDCs; LILRA4+)
- plate- lets (PPBP+)
- hemopoietic stem cells (HSCs; CYTL1+GATA2+).
其umap可视化如下所示:
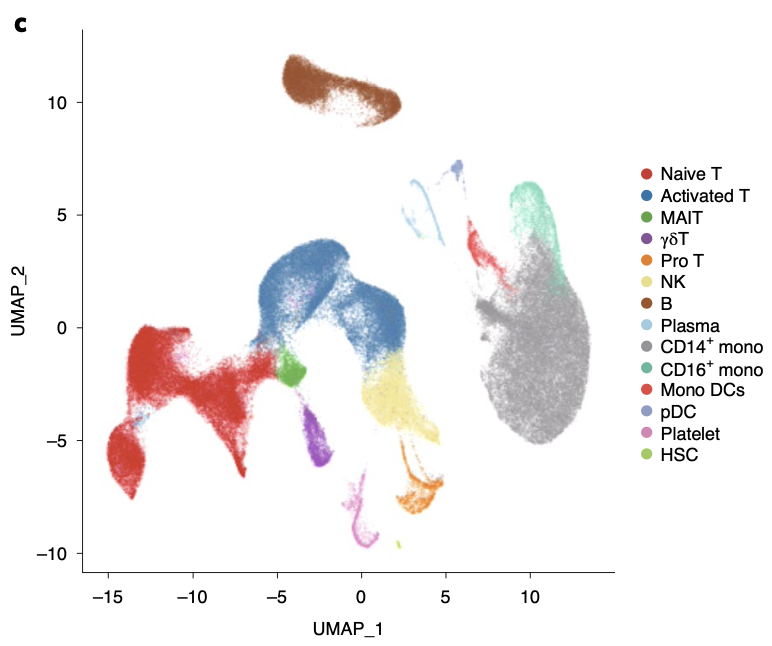
这个配色其实不容易区分,虽然色彩很饱满。每个细胞亚群各自标志性基因的表达量展示,文章也做的很好,这里就不赘述。以前我们做了一个投票:[可视化单细胞亚群的标记基因的5个方法](https://mp.weixin.qq.com/s/enGx9_Sv5wKLdtygL7b4Jw),下面的5个基础函数相信大家都是已经烂熟于心了:
- VlnPlot(pbmc, features = c("MS4A1", "CD79A"))
- FeaturePlot(pbmc, features = c("MS4A1", "CD79A"))
- RidgePlot(pbmc, features = c("MS4A1", "CD79A"), ncol = 1)
- DotPlot(pbmc, features = unique(features)) + RotatedAxis()
- DoHeatmap(subset(pbmc, downsample = 100), features = features, size = 3)
### 核心分析:细胞比例变化和差异分析
单细胞的多组对照设计(例如正常组与给药组)可以为细胞类型水平比较提供以往Bulk RNA-seq分析所不能达到的精度。对此一般有两种进阶分析思路:
- (1)DE(Differential expression)--两组样本的同一细胞类型的基因表达差异分析;
- (2)DA(Differential abundance)--两组样本的同一细胞类型的丰度差异分析
参考:http://bioconductor.org/books/release/OSCA/overview.html
这个文章《Single-cell landscape of immunological responses in patients with COVID-19》的figures2和figures3就分别对应细胞比例差异分析和基因表达量差异分析。
### 各个亚群细分
#### 首先是NK,CD4和CD8细分
第二层次降维聚类分群是:
- 6 subtypes of CD4+ T cells (*CD3E*+*CD4*+),
- 3 subtypes of CD8+ T cells (*CD3E*+*CD8A*+)
- 3 subtypes of NKT cells (*CD3E*+*CD4*–*CD8A*–*TYROBP*+).
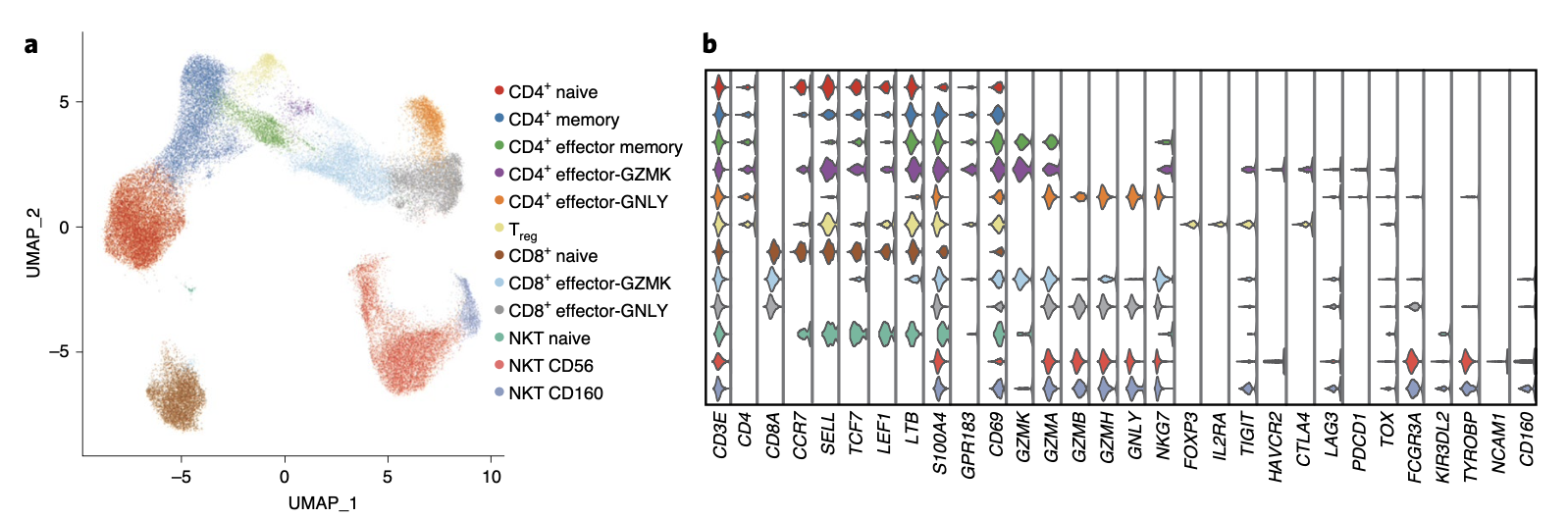
如上图所示,出现了第三层次降维聚类分群:
- naive CD4+ (CD4+ naive) T cell (CCR7+SELL+)
- memory CD4+ (CD4+ memory) T cell (S100A4+GPR183+)
- effector memory CD4+ (CD4+ effector memory) T cell (S100A4+GPR183+GZMA+)
- regulatory T (Treg) cell (FOXP3+IL2RA+)
- naive CD8+ (CD8+ naive) T cell subset (CCR7+SELL+)
- effector CD8+ T cell subsets (CD8+ effector-GZMK and CD8+ effector-GNLY),
- naive NKT (NKT naive) cells (CCR7+SELL+),
- CD56+ NKT (NKT CD56) cells
- CD160+ NKT (NKT CD160) cells
有了这些细分的生物学功能亚群,然后又可以进行核心分析:细胞比例变化和差异分析
单细胞的多组对照设计(例如正常组与给药组)可以为细胞类型水平比较提供以往Bulk RNA-seq分析所不能达到的精度。对此一般有两种进阶分析思路:
- (1)DE(Differential expression)--两组样本的同一细胞类型的基因表达差异分析;
- (2)DA(Differential abundance)--两组样本的同一细胞类型的丰度差异分析
参考:http://bioconductor.org/books/release/OSCA/overview.html
### 划重点:B细胞的细分亚群
如下所示:
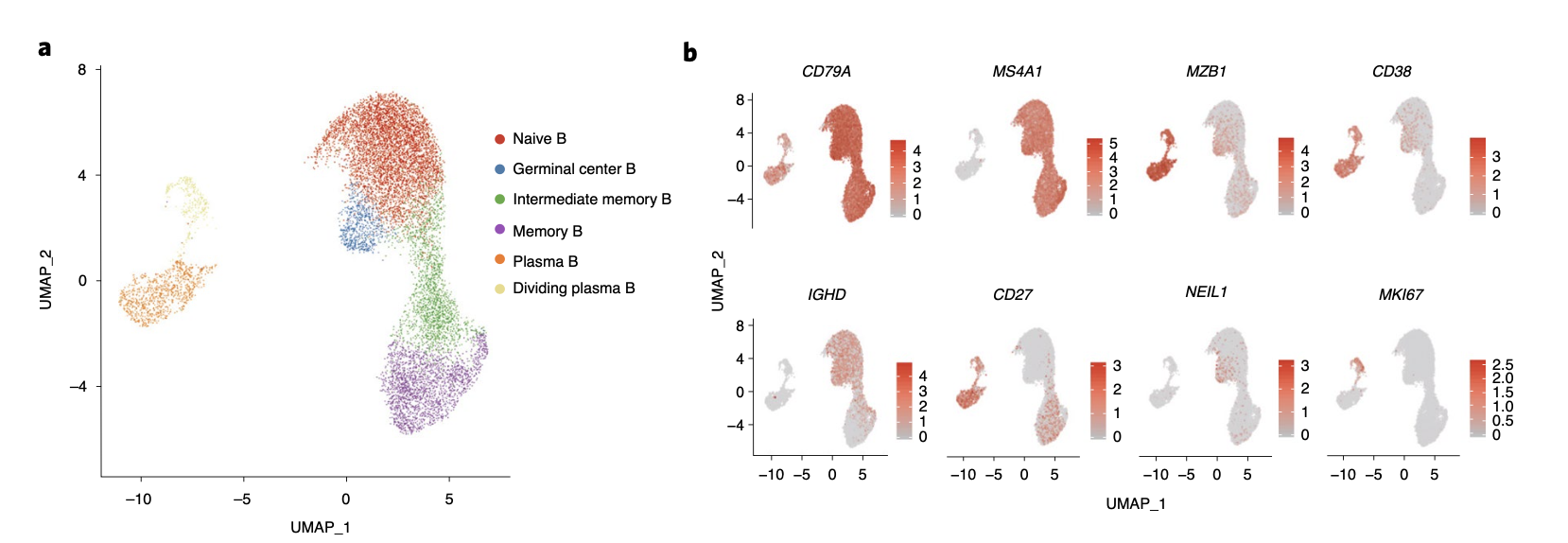
主要是6大亚群:
- one naive B subset (MS4A1+IGHD+)
- one memory B subset (MS4A1+CD27+)
- one intermediate transi- tion memory B subset (intermediate memory B; IGHD+CD27+)
- one germinal center B subset (MS4A1+NEIL1+)
- two plasma subsets plasma B (MZB1+CD38+)
- dividing plasma B (MZB1+ CD38+MKI67+).
当然了,每个亚群各自特异性基因表达量也可视化成功。
更有意思的是,在群里跟大家交流后,提问的小伙伴居然恍然大悟,原来是他自己本来就是看过这个文献。但最开始仅仅是在朋友圈浏览了中文介绍,自己也是下载了原文pdf也是匆匆一瞥。
### 问题在于
没有记录笔记,其实这样的文献对我们的价值并不大,我们能用到的就是降维聚类分群和各个细分亚群标记基因啊!整理起来也非常容易,但是做过就是做过,比匆匆一瞥好太多了。