数据挖掘的本质是把基因的数量搞小,而数据挖掘课题的开启核心就是分组,你可以根据容易基因的高低表达量或者甲基化与否,突变与否来把病人分组, 也可以根据各自生物学功能概念把病人分组。只要分组合理,就可以走差异分析,然后生存分析等等,把基因数量弄小,课题就结束了。
差异分析相信大家都不陌生了,基本上看我六年前的表达芯片的公共数据库挖掘系列推文即可;
- 解读GEO数据存放规律及下载,一文就够
- 解读SRA数据库规律一文就够
- 从GEO数据库下载得到表达矩阵 一文就够
- GSEA分析一文就够(单机版+R语言版)
- 根据分组信息做差异分析- 这个一文不够的
- 差异分析得到的结果注释一文就够
所谓的多组学联合,对普通人来说,不外乎如此。
更糟糕的是,绝大部分人也不会有多组学意识,仍然是根据传统分组, 比如病人年龄分组, 族群分组,比如下面的文章:
Differences in gene-expression profiles in breast cancer between African and European-ancestry women
Jie Ping, Xingyi Guo, Fei Ye, Jirong Long, Loren Lipworth, Qiuyin Cai, William Blot, Xiao-Ou Shu, Wei Zheng
Carcinogenesis, Volume 41, Issue 7, July 2020, Pages 887–893, https://doi.org/10.1093/carcin/bgaa035
Published: 08 April 2020
数据来源于一个 Southern Community Cohort Study (SCCS) 团体,主要是对比 African American (AA) 和 European American (EA) 的 乳腺癌患者差异差异,是转录组测序, 260 AA and 155 EA 对比找差异,然后去TCGA数据库的 180 AA and 838 EA 验证。
- 19 065 genes (16 586 protein-coding and 2479 lincRNAs)
- 2001 (10.5%) were differentially expressed in EA and AA at a nominal P value < 0.05,
- among which 59 genes (54 protein-coding genes and 5 lincRNAs) reached an FDR-adjusted P value < 0.01
- 31 genes expressed significantly higher in AA than EA women,
- while the remaining 28 genes expressed significantly higher in EA than AA women
这59个基因在两个转录组队列的表达量如下所示:
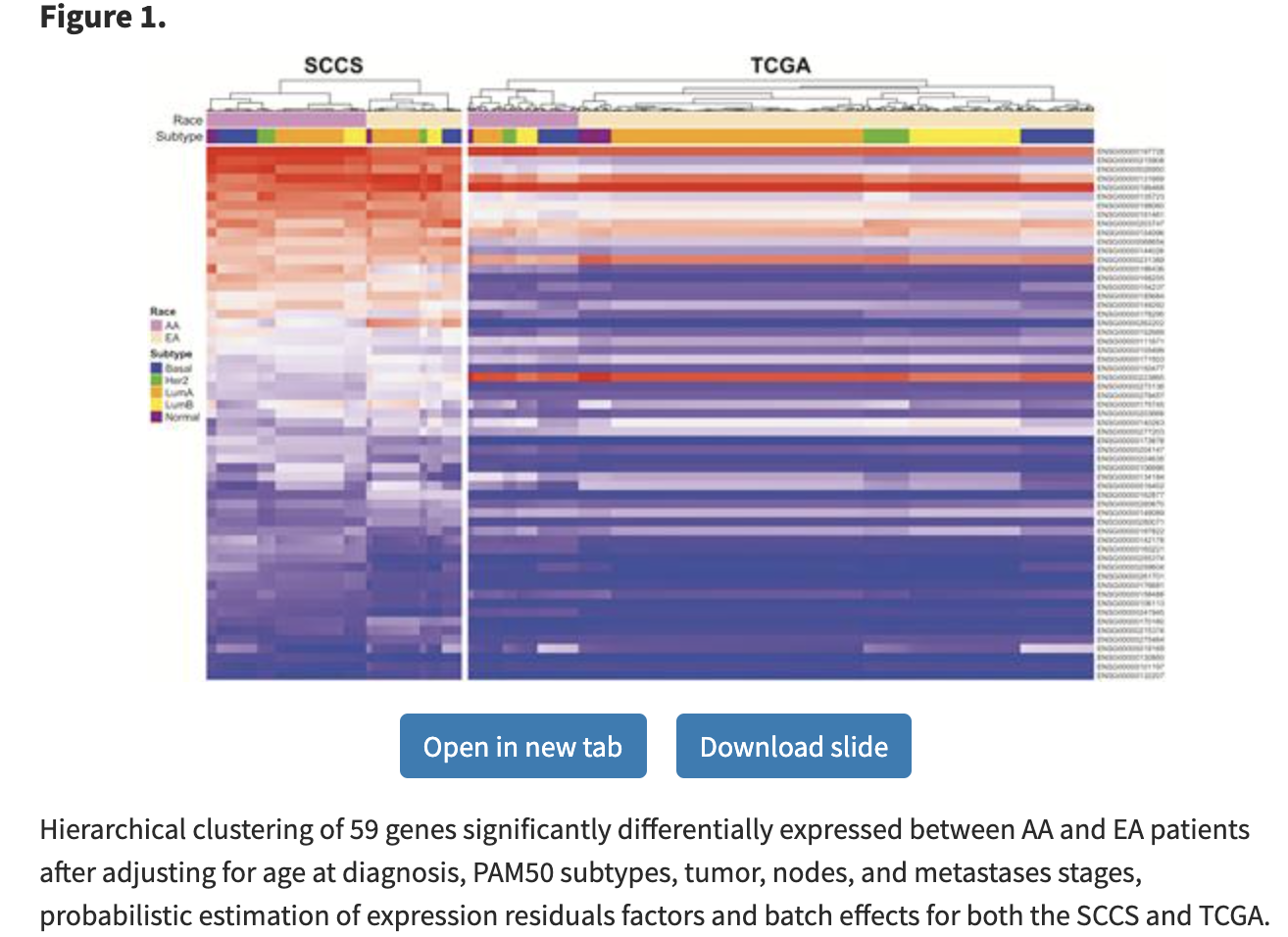
还做了生存分析:
- 10 of the 59 genes were associated with overall survival in AA but not in EA,
- while 7 genes were associated with overall survival in EA but not in AA.
我在生信技能树多次分享过生存分析的细节;
- 人人都可以学会生存分析(学徒数据挖掘)
- 学徒数据挖掘之谁说生存分析一定要按照表达量中位值或者平均值分组呢?
- 基因表达量高低分组的cox和连续变量cox回归计算的HR值差异太大?
- 学徒作业-两个基因突变联合看生存效应
- TCGA数据库里面你的基因生存分析不显著那就TMA吧
- 对“不同数据来源的生存分析比较”的补充说明
- 批量cox生存分析结果也可以火山图可视化
- 既然可以看感兴趣基因的生存情况,当然就可以批量做完全部基因的生存分析
- 多测试几个数据集生存效应应该是可以找到统计学显著的!
- 我不相信kmplot这个网页工具的结果(生存分析免费做)
- 为什么不用TCGA数据库来看感兴趣基因的生存情况
- 200块的代码我的学徒免费送给你,GSVA和生存分析
- 集思广益-生存分析可以随心所欲根据表达量分组吗
- 生存分析时间点问题
- 寻找生存分析的最佳基因表达分组阈值
- apply家族函数和for循环还是有区别的(批量生存分析出图bug)
- TCGA数据库生存分析的网页工具哪家强
- KM生存曲线经logRNA检验后也可以计算HR值
生存分析是目前肿瘤等疾病研究领域的点睛之笔!我们在《生信技能树》b站有两个生存分析免费视频课程, 不知道你能否找到?
其实有了基因集,常规分析都可以走一波!