一个学徒跟着我做了七十多个转录组项目了,但是一直不能理解,凭什么这样的高通量筛选就能定位到具体的一两个基因。
为了帮助他理解生物学的混沌思想,我特意给他找了一个与2018年2月发表在CELL杂志的文章《CD10 + GPR77 + Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness》,这个文章不使用单细胞技术,仅仅是凭借一个高通量的表达量芯片,就筛选到了一个全新的细胞亚群。
文章主旨很明确:
To investigate whether the heterogeneous CAFs contribute to chemoresistance, we isolated fibroblasts from seven chemo- resistant breast cancer biopsies and seven chemosensitive ones obtained before neo-adjuvant chemotherapy.
临床纳入了化疗耐受和敏感的两个人群队列各7人,然后获取他们的成纤维细胞后去探究其异质性。
技术也很简单,就是表达量芯片:
we performed mRNA microarray analysis to compare the mRNA expression profiles of CAFs isolated from the primary tumor biopsies of seven sensitive patients and seven resistant ones before neo-adjuvant chemotherapy.
数据集是: GEO:GSE108565
样品信息如下所示:
GSM2905162 R-1: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient1
GSM2905163 R-2: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient2
GSM2905164 R-3: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient3
GSM2905165 R-4: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient4
GSM2905166 R-5: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient5
GSM2905167 R-6: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient6
GSM2905168 R-7: Breast cancer_neo-adjuvant chemotherapy resistant_CAFs_patient7
GSM2905169 S-1: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient1
GSM2905170 S-2: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient2
GSM2905171 S-3: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient3
GSM2905172 S-4: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient4
GSM2905173 S-5: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient5
GSM2905174 S-6: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient6
GSM2905175 S-7: Breast cancer_neo-adjuvant chemotherapy sensitive_CAFs_patient7
很整齐,两个分组,临床纳入了化疗耐受和敏感的两个人群队列各7人,芯片是NimbleGen Homo sapiens Expression Array [100718_HG18_opt_expr] 并不是主流,不过数据处理是OK的,我已经测试过了。常规的差异分析呢,基本上看我六年前的表达芯片的公共数据库挖掘系列推文即可;
- 解读GEO数据存放规律及下载,一文就够
- 解读SRA数据库规律一文就够
- 从GEO数据库下载得到表达矩阵 一文就够
- GSEA分析一文就够(单机版+R语言版)
- 根据分组信息做差异分析- 这个一文不够的
- 差异分析得到的结果注释一文就够
作者发现差异并不是想象的那些:
Although CAFs from chemosensitive and resistant patients exhibited distinctive mRNA signatures (Figure 1C), conventional fibroblast markers, including a-SMA, PDGFRb, FAP, FSP1, and collagen I, failed to distinguish them (Table S1).
我觉得这句话好奇怪,既然对化疗耐受和敏感的两个人群队列都是看成纤维细胞的表达量,那么传统的成纤维细胞的标记基因理论上是不会有表达量差异啊。
不过作者强行讲解他的生物学故事,因为他就是要搞清楚化疗耐受和敏感的两个人群队列的成纤维细胞的表达量差异情况:
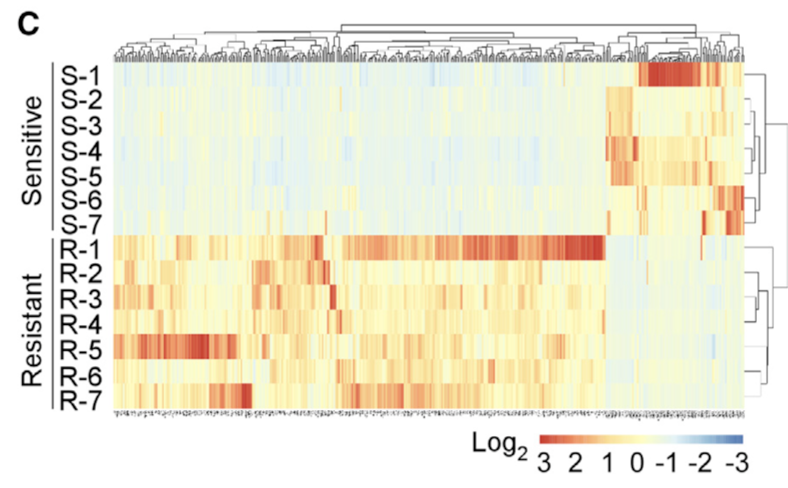
(C) Heatmap representing differential expressed genes (fold change > 3) of the CAFs isolated from the biopsies of seven chemoresistant and seven chemo- sensitive patients before neo-adjuvant chemotherapy.所以检查cell-surface markers
这个时候大家一定会好奇,只要是差异分析,如果实验设计合理,肯定是有成百上千个基因表达量变化,而且是具有统计学显著的,凭什么作者这个时候要检测cell-surface markers呢?这就是传说中的“生物学背景知识”啦,
We then searched for cell-surface markers to identify these CAFs by evaluating differentially expressed mRNAs that encode membrane proteins and found four of them upregulated in the CAFs of the resistant tumors versus those derived from the sensitive ones. Among them, upregulation of CD10 and GPR77 in the CAFs from chemoresistant tumors was validated by qRT-PCR in another cohort of 24 patients (Figure 1D).
当然了,仅仅是高通量筛选,通常是被认为可靠性欠缺,所以作者这个时候使用 qRT-PCR 的实验验证一波:
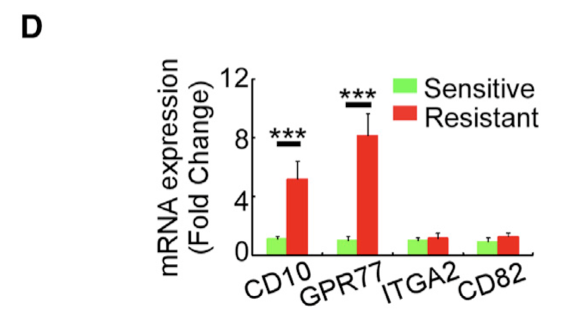
(D) The differential mRNA expression for cell-surface proteins was validated by qRT-PCR in CAFs isolated from pre-treatment breast cancer biopsies of chemosensitive (n = 13) and chemoresistant (n = 11) patients.生物学功能数据库GSEA方法富集
这方面教程以及不计其数了,其实GSEA方法富集也会面临同样的问题,因为生物学功能数据库的通路也是好几万的数量,只要是你去分析,一定能找到统计学显著的。
这个时候,仍然是传说中的“生物学背景知识”为你撑腰。
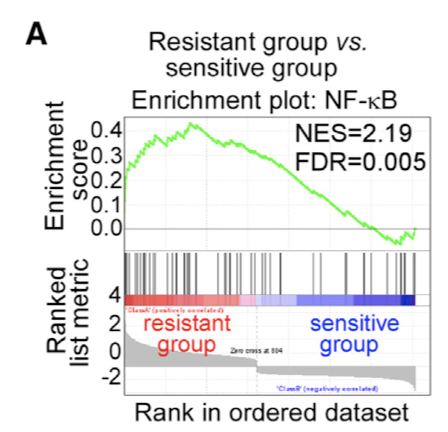
(A) GSEA analysis revealed an enrichment of NF-kB target genes in the CAFs from chemoresistant breast cancer samples. The heatmap of differential expression profiles was illustrated in Figure 1C.学徒作业
在 Table S1. The mRNA expression profiles of CAFs, Related to Figure 1 可以看到作者得到的差异基因列表:
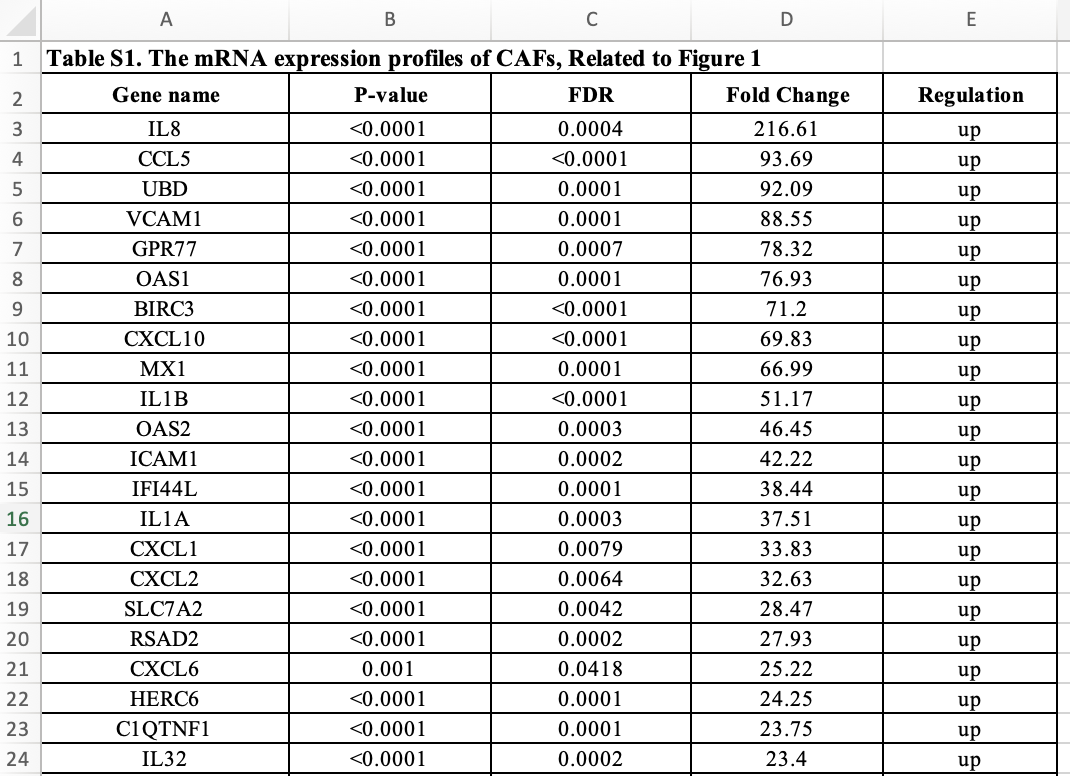
我希望你处理数据集: GEO:GSE108565,检查这个结果。思考题
在没有单细胞技术的加持情况下,细胞亚群的命名问题,我看到了另外一个文献:
CD63+ Cancer-Associated Fibroblasts Confer Tamoxifen Resistance to Breast Cancer Cells through Exosomal miR-22. Adv Sci (Weinh). 2020 Sep 24;7(21):2002518.文末友情推荐
- 学徒培养2021名额开放申请
- 老板,请为我配备一个懂生信的师兄
- 你以为GEO只是挖挖就完了吗