很早以前我就在《生信技能树》的推文:新的ngs流程该如何学习(以CUT&Tag 数据处理为例子),提到了我自己是不太可能去把所有的ngs流程全部录制视频的,只能说是更好的传达学习方法给到大家。
其实如果你看过我表观组学系列,比如《ChIP-seq数据分析》 和 《ATAC-seq数据分析》 就会知道这些技术都可以被单细胞化, 如果你具备比较好的背景知识,理论上是可以自己根据文档把它们对应的单细胞水平的数据分析摸索成功。那就作为学徒作业吧,摸索scChIPseq流程!
文章是:High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 2019 Jun;51(6):1060-1066. PMID: 31152164
数据在:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE117309
GSM3290887 H3K4me3_scChIPseq_Jurkat-Ramos
GSM3290888 H3K27me3_scChIPseq_Jurkat-Ramos
GSM3290889 H3K27me3_scChIPseq_HBCx-95
GSM3290890 H3K27me3_scChIPseq_HBCx-95-CapaR
GSM3290891 H3K27me3_scChIPseq_HBCx-22
GSM3290892 H3K27me3_scChIPseq_HBCx-22-TamR
# 下面的4个单细胞都是10x技术的
GSM3290893 H3K27me3_scRNAseq_HBCx-95
GSM3290894 H3K27me3_scRNAseq_HBCx-95-CapaR
GSM3290895 H3K27me3_scRNAseq_HBCx-22
GSM3290896 H3K27me3_scRNAseq_HBCx-22-TamR
可以看到,区分T和B细胞亚群的效果非常好!
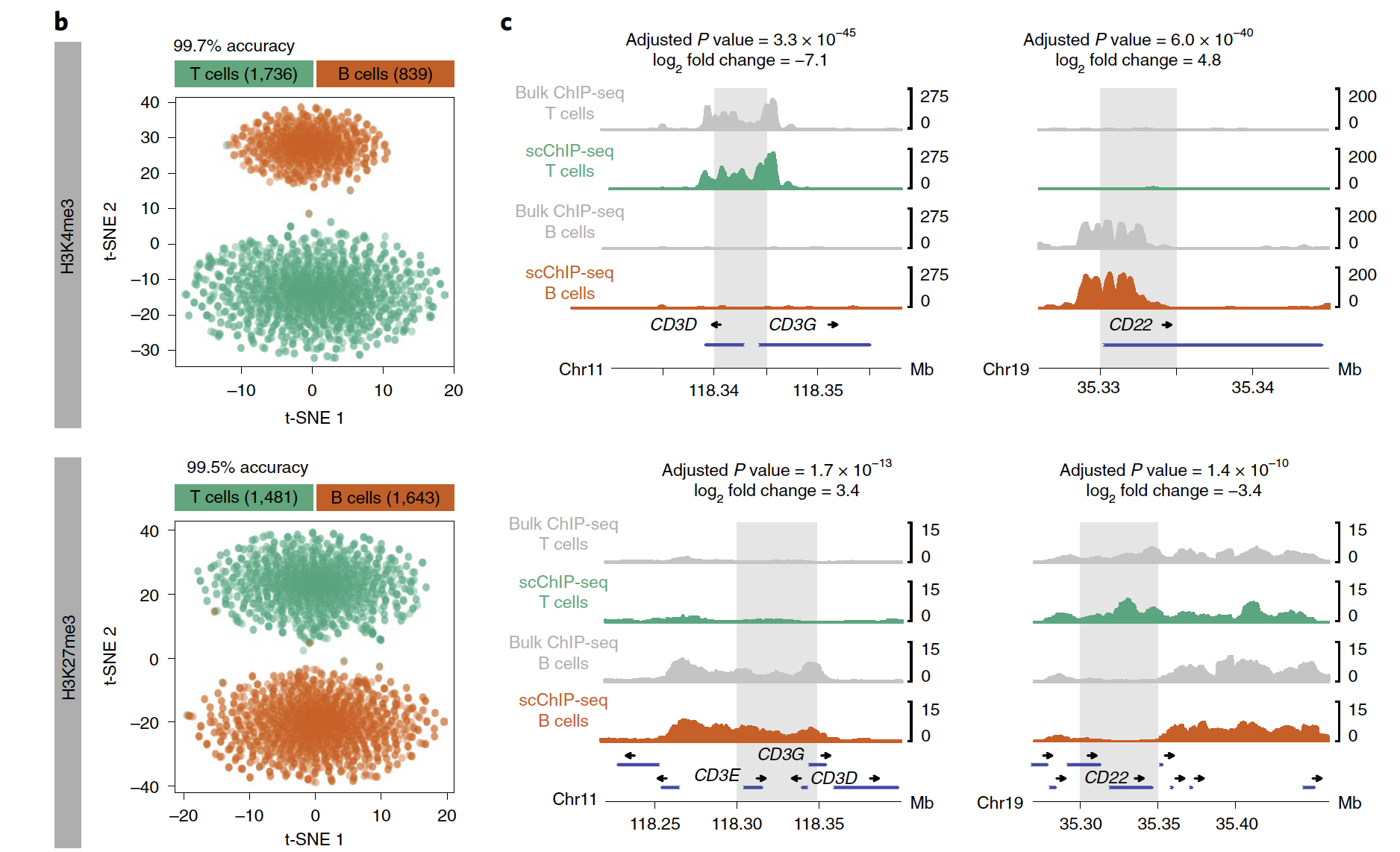
两种组蛋白修饰情况:
- H3K4me3: n = 1,736 T cells and 839 B cells;
- H3K27me3: n = 1,481 T cells and 1,643 B cells
这两个两种组蛋白修饰的生物学意义完全不一样哦:
数据分析方面也是拿到矩阵,主要是Bowtie v.1.2.2比对到mm10和 hg38两个 参考基因组,去除PCR重复,:
- We generated a coverage matrix and metrics from these de-duplicated reads, referred to as ‘unique reads’ in the text.
- For each cell, reads were binned in non-overlapping 50 kb bins for H3K27me3, known to accumulate over broad genomic regions, and 5 kb genomic bins for H3K4me3,
- known to accumulate in narrow peaks around TSSs, spanning the genome, to generate an n X m coverage matrix with n barcodes and m genomic bins.
如果你没有单细胞转录组认知,需要先看看基础10讲:
- 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
- 09.单细胞转录组数据处理之细胞亚群比例比较
最基础的往往是降维聚类分群,参考前面的例子:人人都能学会的单细胞聚类分群注释