我们非常强调进入一个领域需要读综述来获取基本认知,尤其是单细胞,我们在《单细胞天地》公众号给大家精选了2017-2020的4篇综述:
2017年7月的 Identifying cell populations with scRNASeq
| https://www.ncbi.nlm.nih.gov/pubmed/28712804
2018年2月的 Single-cell RNA sequencing: Technical advancements and biological applications
| https://www.ncbi.nlm.nih.gov/pubmed/28754496
2019年9月的 Current best practices in single-cell RNA-seq analysis: a tutorial
| https://www.ncbi.nlm.nih.gov/pubmed/31217225
2020年3月的 Tools for the analysis of high-dimensional single-cell RNA sequencing data
| https://www.ncbi.nlm.nih.gov/pubmed/32221477
大家可以自行前往《单细胞天地》公众号寻找其对应的中文翻译整理版本,现在是2021了,所以再加一个综述:《Critical downstream analysis steps for single-cell RNA sequencing data》,Briefings in Bioinformatics, , https://doi.org/10.1093/bib/bbab105
- PIPELINE FOR ANALYSING scRNA-seq TECHNOLOGY
- QUALITY CONTROL AND DIFFERENTIAL GENE EXPRESSION ANALYSIS
- CLUSTERING
- TRAJECTORY INFERENCE
- CELL-TYPE ANNOTATION
- INTEGRATING DATASETS
主要是这个综述罗列了大量的工具, Table 1. Clustering methods for single-cell RNA sequencing data
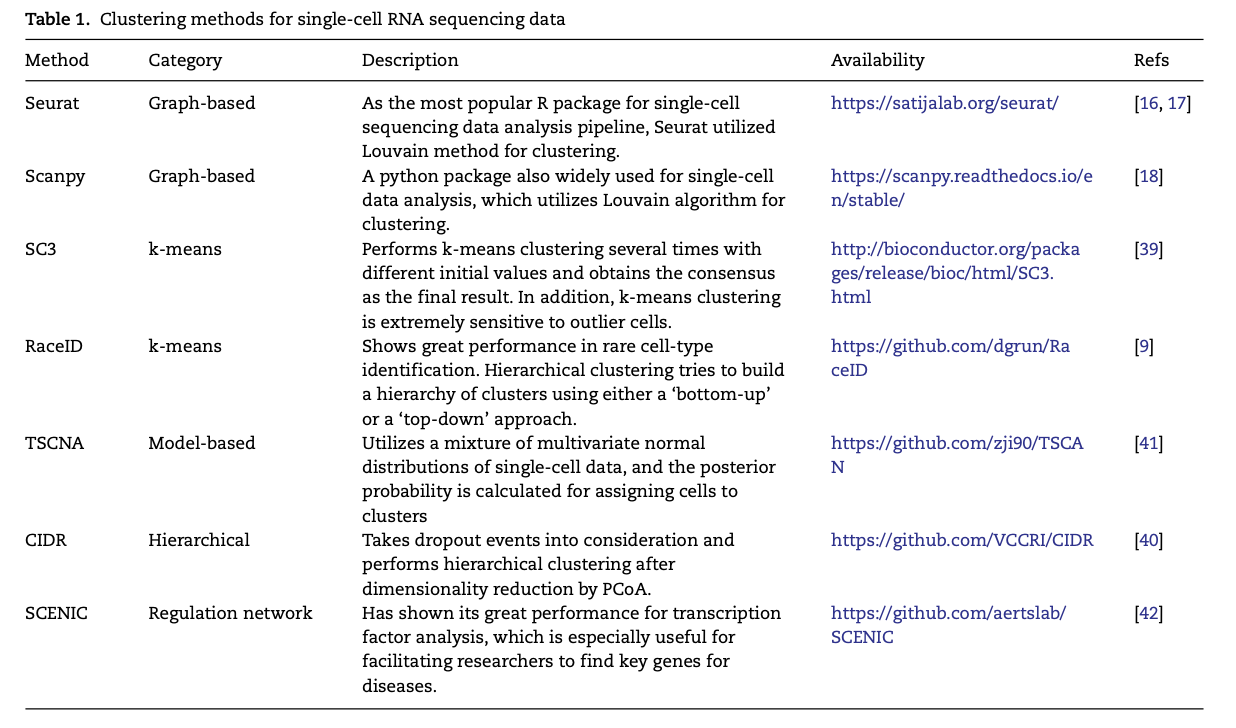
还有
- Table 2. Trajectory inference methods for single-cell RNA sequencing data
- Table 3. Cell-type annotation methods for single-cell RNA sequencing data
- Table 4. Popular data resources for cell-type annotation
- Table 5. Integrating datasets methods for single-cell RNA sequencing data
挺容易看懂的,今年入坑单细胞的,可以优先看看这个!其实这样的基础认知,也可以看基础10讲:
- 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
- 09.单细胞转录组数据处理之细胞亚群比例比较
最基础的往往是降维聚类分群,参考前面的例子:人人都能学会的单细胞聚类分群注释