最近看到一个文章,标题是:《Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis》,发表于February 5, 2018 https://doi.org/10.3892/ijo.2018.4265 实验设计超级简单:
-
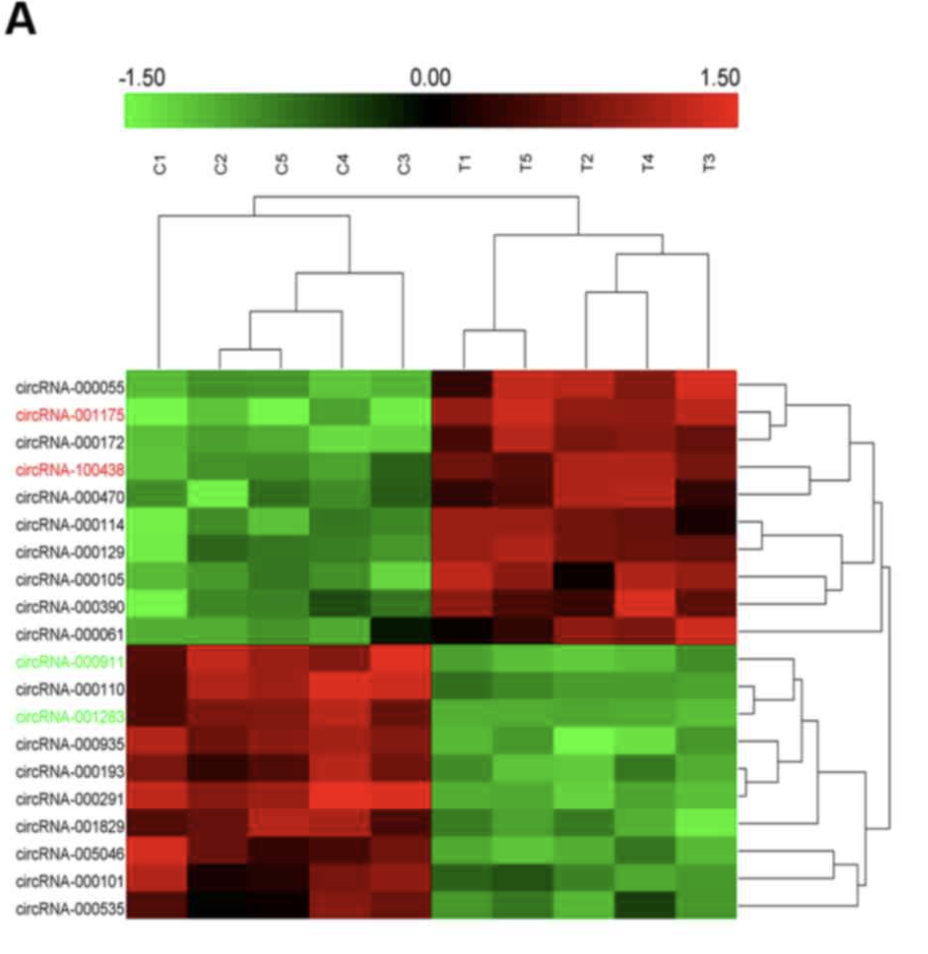
研究者首先在真实的病人队列做了5 tissues samples and control non-tumor tissue,差异分析结果是:A total of 716 circRNAs that were differently expressed by >1.5-fold between the breast cancer tissues and the normal tissues were identified.
-
然后从 Chinese Academy of Sciences (Shanghai, China).获得了 这些细胞系:MCF-7, MDA-MB-231, MDA-MB-468, MDA-MB-453, SKBR-3 and T47D, and one normal breast epithelial cell line (MCF-10A) .
本来呢,我非常开心,看到了文章里面提到了数据可以下载哦, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97332 ,所以想安排给学徒们做练习题。毕竟也是一个表达矩阵, 可以做差异分析等等:
但是我进入GEO界面查看,诡异的事情发生了:
GSM2561829 hepar normal tissue circRNA 1
GSM2561830 hepar normal tissue circRNA 2
GSM2561831 hepar normal tissue circRNA 3
GSM2561832 hepar normal tissue circRNA 4
GSM2561833 hepar normal tissue circRNA 5
GSM2561834 hepar normal tissue circRNA 6
GSM2561835 hepar normal tissue circRNA 7
GSM2561836 hepatocellular carcinoma(HCC) tumor tissue circRNA 1
GSM2561837 hepatocellular carcinoma(HCC) tumor tissue circRNA 2
GSM2561838 hepatocellular carcinoma(HCC) tumor tissue circRNA 3
GSM2561839 hepatocellular carcinoma(HCC) tumor tissue circRNA 4
GSM2561840 hepatocellular carcinoma(HCC) tumor tissue circRNA 5
GSM2561841 hepatocellular carcinoma(HCC) tumor tissue circRNA 6
GSM2561842 hepatocellular carcinoma(HCC) tumor tissue circRNA 7
怎么好好的乳腺癌研究,变成了肝癌呢?我看了看这个数据集, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE97332 ,跳转的文献是:Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology 2017 Oct;66(4):1151-1164. PMID: 28520103
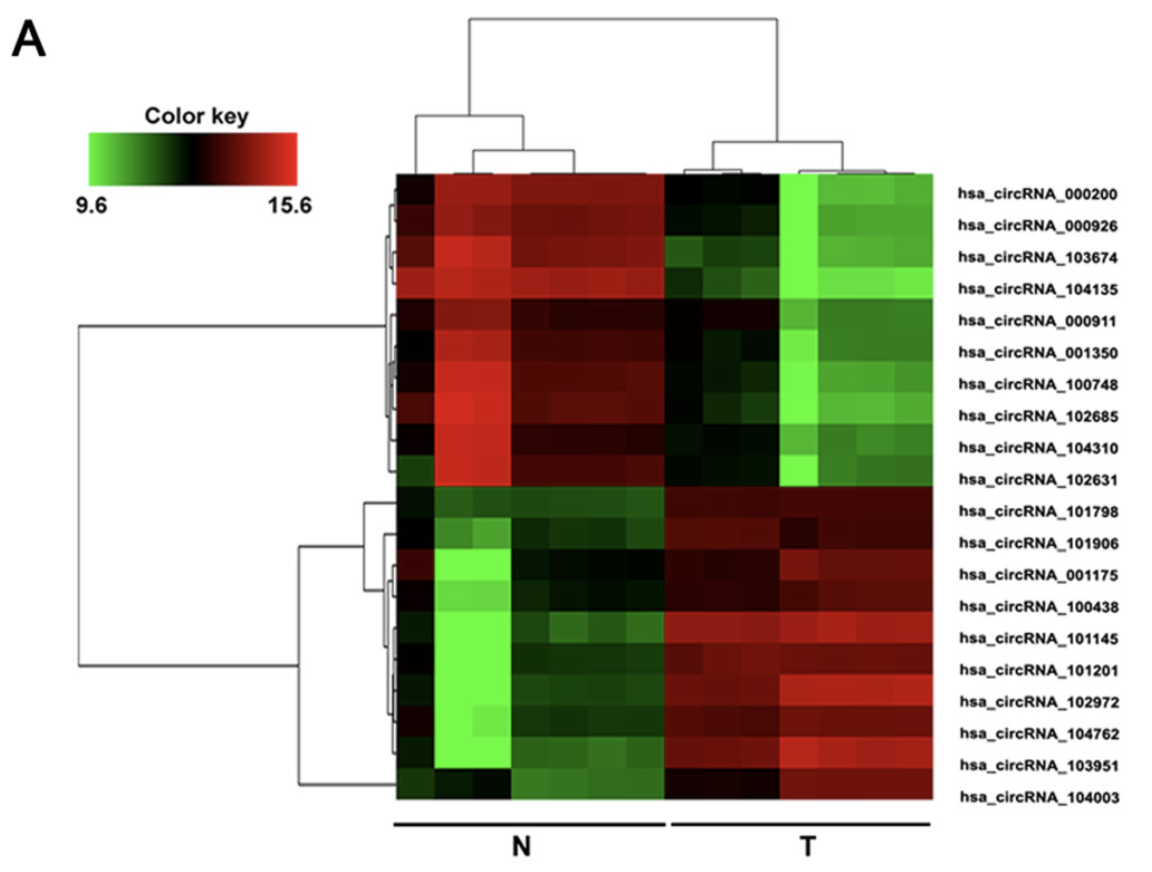
(A) The heat map showed the top 10 most increased and decreased circRNAs in HCC tissues as compared to that in the matched nontumor tissues analyzed by circRNAs Arraystar Chip.
我就怕自己眼瞎,认真看了好几遍哦,这个基因芯片是: The circRNAs chip (ArrayStar Human circRNAs chip; ArrayStar, Rockville, MD, USA) containing 5,639 probes specific for human circular RNAs splicing sites was used.
The circRNA microarray process was performed by KangChen Biotech, Inc. (Shanghai, China).
这是一个有故事的公司!