看到2020年2月发表在nature cancer这个新杂志的文章《Single-cell analyses reveal increased intratumoral heterogeneity after the onset of therapy resistance in small-cell lung cancer》链接是:https://www.nature.com/articles/s43018-019-0020-z 里面有普通的18个样品的bulk转录组数据以及25个单细胞转录组数据。

主要是 (CTC)-derived xenografts (CDXs) ,听说过是技术难度很高!
首先是突变全景图很突兀,也很诡异
如下:
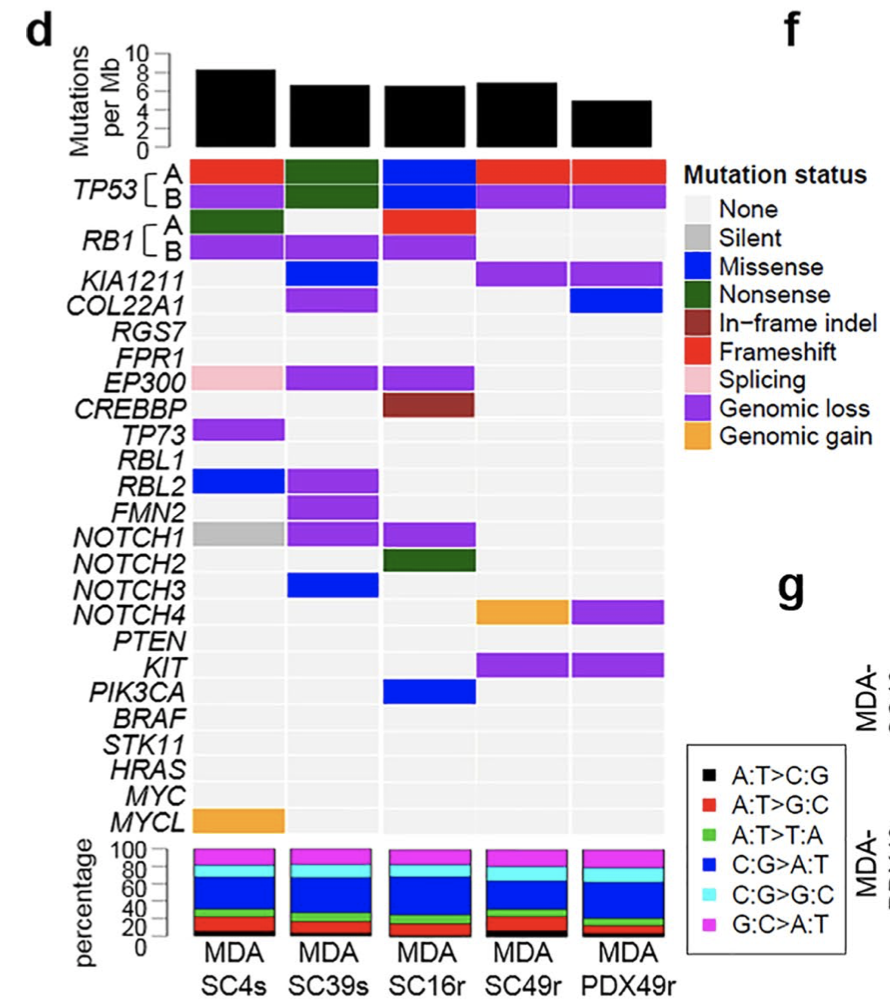
全文反正也没有提到如何测序,是WGS,WES还是说仅仅是 panel,没有突变数据分析的描述,仅仅是提到了BWA和VARSCAN软件而已,也不上传数据。这样的话,我们只能是假设研究者没有造假咯!
反正没有数据可以下载,而且作者描述的也超级简陋,做啥图表复现都不可能额!
单细胞呢,好像缺一个样品!!
单细胞的实验环节,正文里面说的倒是很清楚,是目前主流的10X商业化仪器的数据 :
Sorted cells were washed once with 0.04% bovine serum albumin in phosphate-buffered saline and counted on a Countess II automated cell counter (Thermo Fisher Scientific). A total of 12,200 cells were loaded per lane on the 10× Chromium platform and processed for complementary DNA synthesis and library preparation, per the manufacturer’s protocol using version 2 chemistry. Complementary DNA and libraries were checked for quality on an Agilent 4200 TapeStation and quantified by KAPA quantitative PCR before sequencing on a single lane of a HiSeq 4000 (Illumina).
数据分析细节如下:
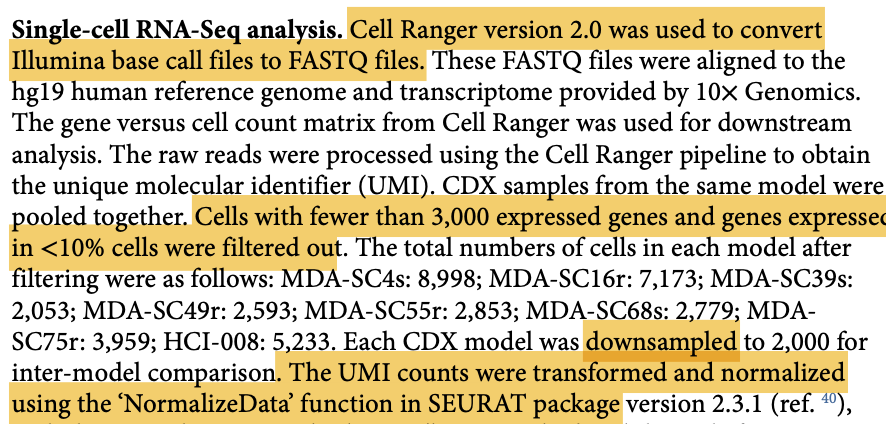
完全没办法接受研究者的阈值,居然是表达的基因数量少于3000的细胞直接就过滤了!!!这个是10X商业化仪器的数据 啊~~~
而且明明是8个单细胞转录组的 CDXs 数据 ,正文也写了; t-SNE analysis of eight CDXs. 结论是: Cells from each CDX were more similar to themselves than to other models. 但是上传的数据呢,缺失那个 HCI开头的样品:
- https://www.ncbi.nlm.nih.gov/bioproject/PRJNA575243
- https://www.ebi.ac.uk/ena/browser/view/PRJNA575243
反正我找了很久,都是看不到的!
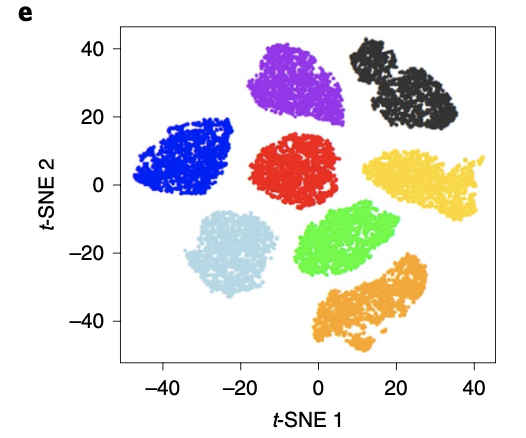
这8个单细胞转录组数据的独立聚类分群如下:
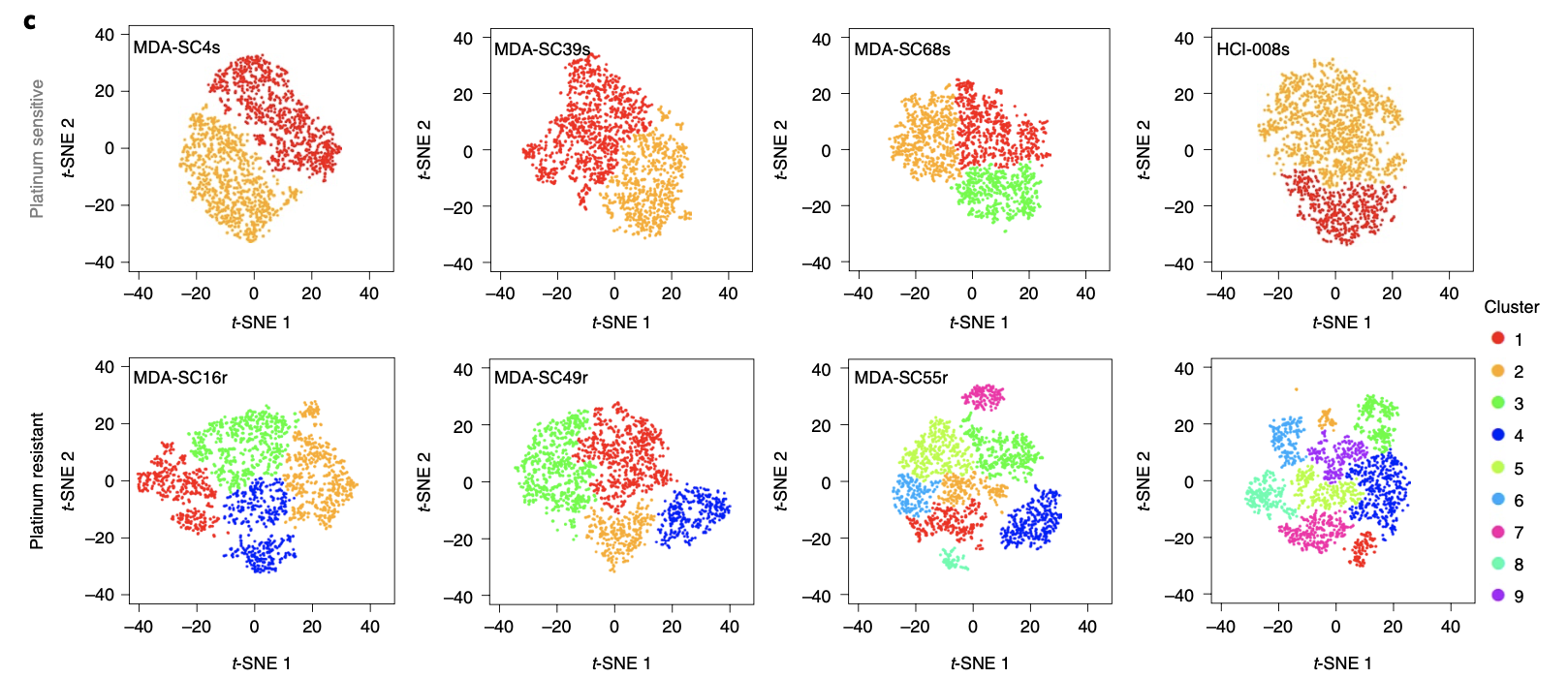
这些分析都超级简单,基本上单按照细胞基础10讲 - 01. 上游分析流程
- 02.课题多少个样品,测序数据量如何
- 03. 过滤不合格细胞和基因(数据质控很重要)
- 04. 过滤线粒体核糖体基因
- 05. 去除细胞效应和基因效应
- 06.单细胞转录组数据的降维聚类分群
- 07.单细胞转录组数据处理之细胞亚群注释
- 08.把拿到的亚群进行更细致的分群
- 09.单细胞转录组数据处理之细胞亚群比例比较
一步步分析即可!虽然原始数据可以在EBI下载原始数据,但是缺关键信息
根据 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138267 可以拿到ebi数据库链接,链接是:https://www.ebi.ac.uk/ena/browser/view/PRJNA575243
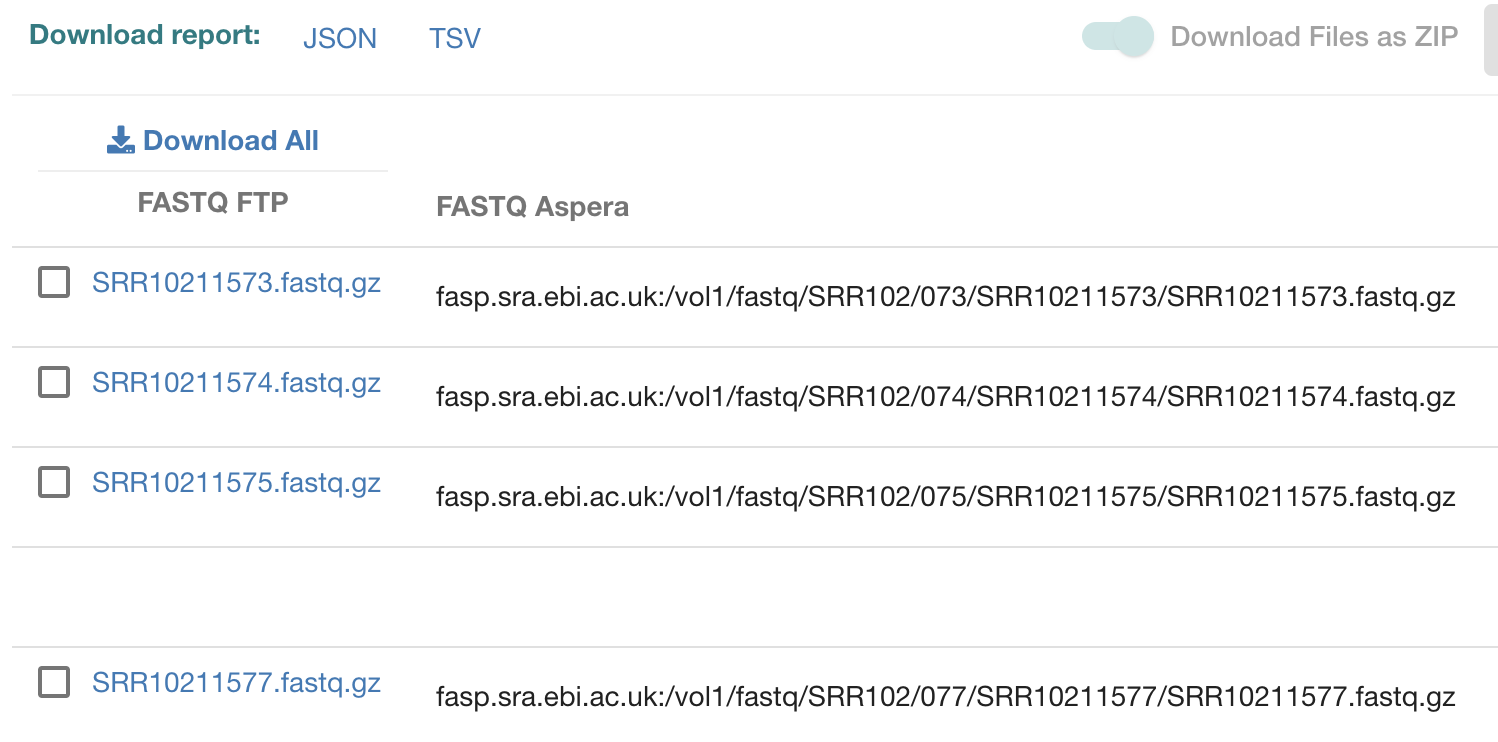
如下所示,每个单细胞样品,都仅仅是提供了一个fastq文件:
其实前面我们提到过,10X的单细胞转录组原始数据的话, 比较特殊,它的测序文库中包括index、barcode、UMI和测序reads。 - 首先,1-26个cycle就是测序得到了26个碱基,先是16个Barcode碱基,然后是10个UMI碱基;
- 然后,27-34这8个cycle得到了8个碱基,就是i7的sample index;
- 最后35-132个cycle得到了98个碱基,就是转录本reads,需要拿去做比对的!
作者仅仅是提供一个fastq文件,基本上可以定论,这样的数据没意义了。但是为了让我们的结论更保险,我还是亲自下载看看,可以参考:使用ebi数据库直接下载fastq测序数据 , 需要自行配置好,然后去EBI里面搜索到的 fq.txt 路径文件。这里演示一个数据的下载即可:conda activate download dsa=$HOME/miniconda3/envs/download/etc/asperaweb_id_dsa.openssh ls -lh $dsa nohup ascp -QT -l 300m -P33001 -i $dsa \ era-fasp@fasp.sra.ebi.ac.uk:/vol1/fastq/SRR102/073/SRR10211573/SRR10211573.fastq.gz ./ &花了半个小时下载了这个接近 20G的数据,看了看前面的一点点:
@SRR10211573.1 K00384:78:HLM37BBXX:3:1101:27630:1261/2 CATTTTGTANTACGGGGATACCTGNGACTGCACCNTTAAAAAATATATTTATCATTTAANTCTTGGGTAANCACACTTCATAACAGAGNAGAGNGANT + A<<-----<#--7--7---77A-7#7JJFAAJ7J#7-FF-<--<--77--77AF<----#-<<----<77#FJ-<-AJ--7--7-7--#-7-<#--#- @SRR10211573.2 K00384:78:HLM37BBXX:3:1101:27813:1261/2 AGTGAATCCNAATACTTACAGCCCNCTGATGTGCNCCAAAGTAAACAAAAACAATGATGNTAATGGACACNCTCTTCAATATACTAGGNGGAGNGGNG + A---<<---#--<A<<F<F-FFF-#7-7---7-<#---7F<-AA<F-<-AAFJF---7-#-AF-7-7-7A#F7-777-----7---<-#-7--#7-#7很正常的转录本reads数据信息,但是缺Barcode文件信息。 假如作者提供的10x数据是合格的,就可以走我们的流程即可,参考我在《生信技能树》的教程:cellranger更新到4啦(全新使用教程)
目前单细胞转录组以10X公司为主流,我们也是在单细胞天地公众号详细介绍了cellranger流程,大家可以自行前往学习,如下:
- 单细胞实战(一)数据下载
- 单细胞实战(二) cell ranger使用前注意事项
- 单细胞实战(三) Cell Ranger使用初探
- 单细胞实战(四) Cell Ranger流程概览
- 单细胞实战(五) 理解cellranger count的结果
但是这个两年前的系列笔记是基于V2,V3版本的cellranger,目前呢它更新到了版本4,建议以我的最新版教程为准,在《生信技能树》的教程:cellranger更新到4啦(全新使用教程)唯一可利用的居然是表达矩阵
作为学徒作业吧,大家进入 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE138267 下载全部的表达矩阵,走一下文章里面的聚类分群看看,重复性如何!
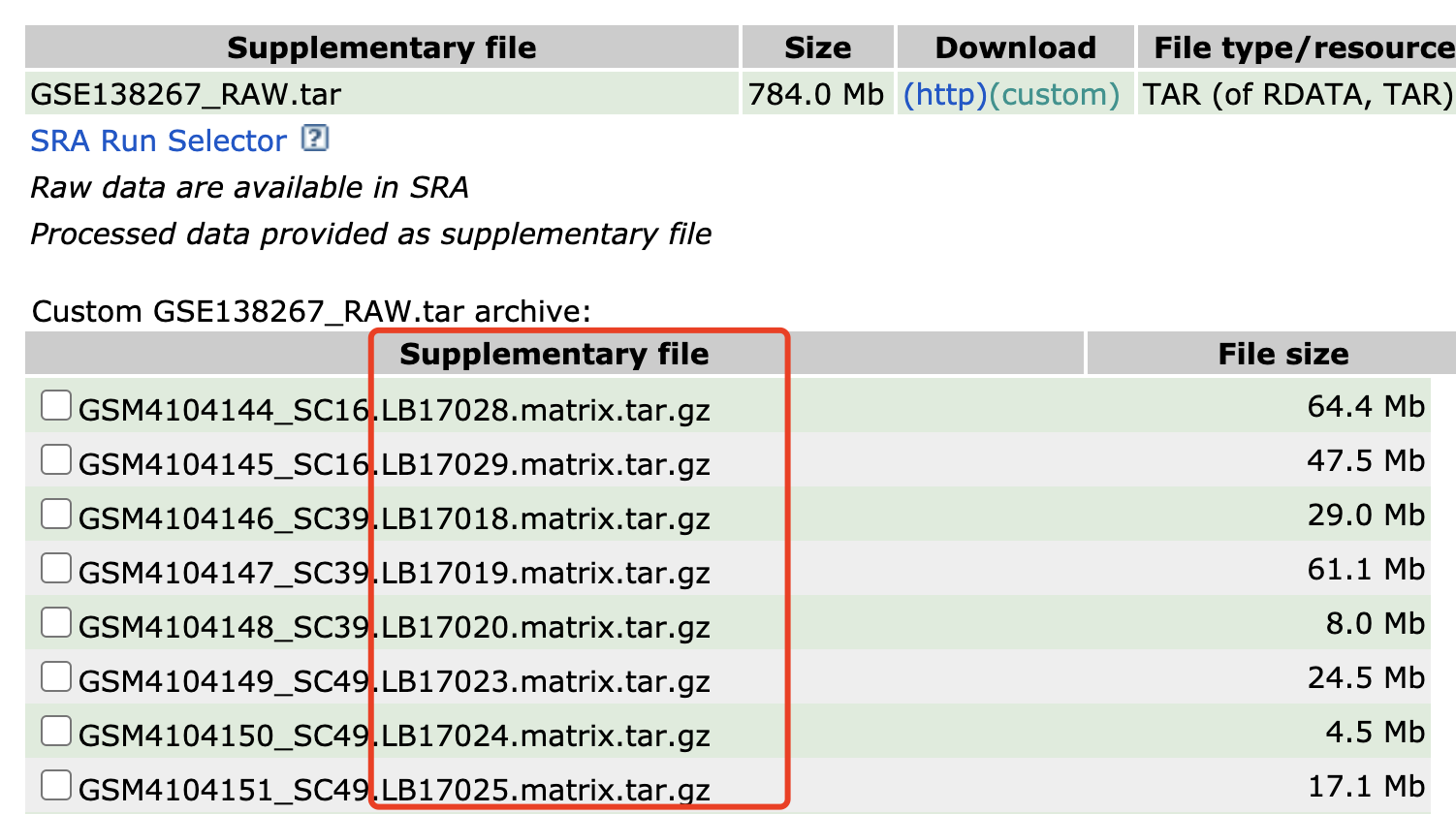
每个样品都是一个独立的压缩包,都是3个文件,如下:
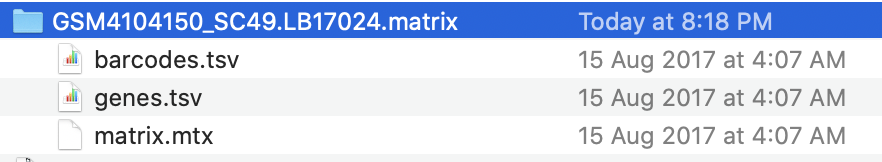
走我们一直强调的10X数据处理全套代码即可。
检查是不是缺失一个样品,是不是表达的基因数量少于3000的细胞之间就过滤了?