文献《Multi-Omics Profiling Reveals Distinct Microenvironment Characterization and Suggests Immune Escape Mechanisms of Triple-Negative Breast Cancer》,研究者把TNBC根据免疫分成3个亚群,然后寻找Potential intrinsic immune escape mechanisms of TNBC,这个过程应用了很多突变位点的量化指标,包括:
- neoantigens,
- cancer testis antigens (CTAs),
- homologous recombination deficiency (HRD) scores,
- intratumoral heterogeneity (ITH)
- TMB
结果如下;
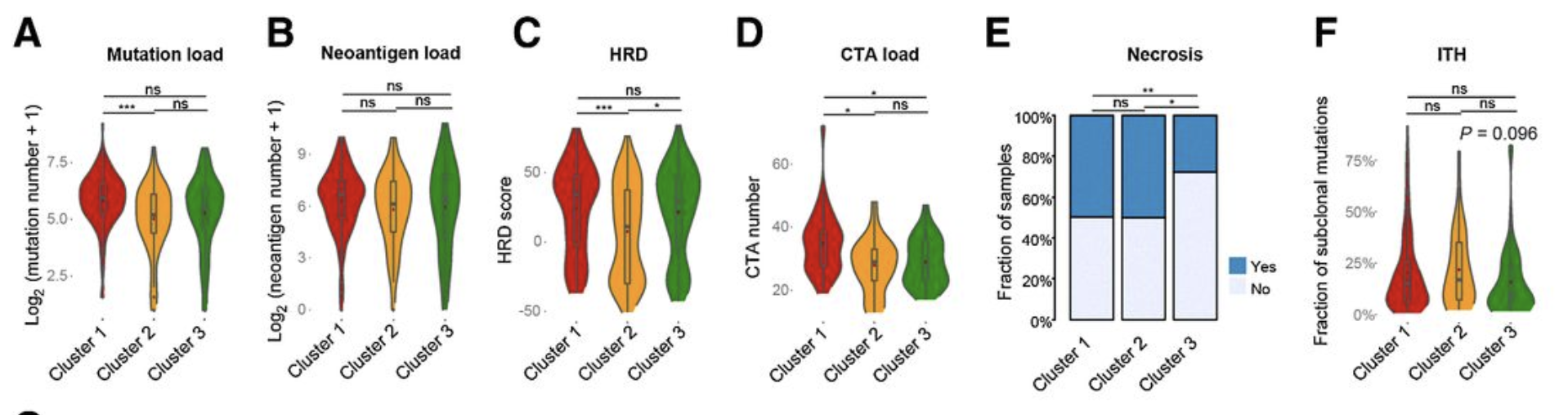
Comparison of mutation loads (A), neoantigen load (B), HRD scores (C), CTA numbers (D), necrosis (E), and ITH scores (F) among the three clusters. In the violin plots, the mean values are plotted as red dots, and the boxplot was drawn inside the violin plot.
计算方法都在附件:https://clincancerres.aacrjournals.org/content/suppl/2019/03/05/1078-0432.CCR-18-3524.DC1
我摘抄了这个英文描述,相信绝大部分人看着都会两眼摸黑:
Calculation of neoantigens
With the WES data (.bam) of paired normal samples from TNBC patients, we first used POLYSOLVER tool (8) to infer the 4-digit HLA genotype for each sample (arguments: Asian 1 hg19 STDFQ 0). Then, neoantigens were predicted using NetMHCpan (v4.0) (9), with the somatic mutation data (.maf) and HLA genotype data as the inputs. Neoantigens derived from protein coding single nucleotide variants (SNV) (Variant_Classification = “Missense_Mutation”, and Variant_Type = ‘‘SNP”) and small insertions and deletions (Indel) (Variant_Classification = “Frame_Shift_Ins’’, ‘‘Frame_Shift_Del’’, ‘‘In_Frame_Ins’’, ‘‘In_Frame_Del’’, and Variant_Type = ‘‘INS”, “DEL”) were predicted separately. Mutations which were predicted to produce peptide with affinity < 500 nM and of which the corresponding gene was expressed greater than Combat value 1 (evaluated based on median expression rather than the specific sample) were chosen as neoantigens. We referred to pVAC-seq (10) and made some modifications based on the features of our dataset to construct this algorithm.
Calculation of cancer testis antigens (CTA)
The CTDatabase (http://www.cta.lncc.br/) was first queried for CTAs. We then calculated the difference in each candidate CTA between the tumor site and the paired normal site; genes whose expression were at least four times higher in the tumor site than the paired normal tissue in at least one patient were selected as TNBC-specific CTAs. In all, a total of 177 CTAs were included in our study. The CTA landscape of TNBC is described in Supplementary Figure 8.
Calculation of homologous recombination deficiency (HRD) scores
The HRD score was calculated as the sum of three scores: allelic imbalance extending to the telomere (NtAI) score, loss of heterozygosity (LOH) score and modified large-scale state transition (LSTm) score. The calculation of these scores was previously described (11). Briefly, the NtAI score was defined as the number of regions with allelic imbalance longer than 11 Mb and extending to one of the subtelomeres but do not crossing the centromere. The LOH score was defined as the number of LOH regions longer than 15 Mb but shorter than the whole chromosome. The LST score was defined as the number of break points between regions longer than 10 Mb after filtering out regions shorter than 3 Mb. In order to diminish effect of ploidy, The LST score was modified using the following formula: LSTm = LST – kP, where P is ploidy, and k is a constant of 15.5.
Estimation of intratumoral heterogeneity (ITH)
ASCAT (12) was used to integrate the copy number data with the data on somatic mutations to estimate the purity and ploidy of each tumor using default parameters. A modified PyClone workflow (13) was then used to estimate the cancer cell fractions of each sample. The fraction of subclonal cancer cells was set as indicators representing the ITH.那么有没有捷径学这些方法呢
当然是有的,在华大和诺禾都工作过了的十多年生信项目经验的讲师手把手小班教学,你值得拥有,详情请点击:肿瘤信息学实战学习班(仅限30人),第二期招生现在(2020-11-26 )开始,同样的名额有限,理论上很快就招满了!
课程详细说明:
一、 变异检测研究-SNP/INDEL
Somatic SNP/INDEL变异是肿瘤基因组学研究的基础。然而受限于肿瘤的异质性与其突变频率的特点,准确地检出这些变异仍然是需要足够的经验的。我们围绕Broad Institute的Somatic SNVs +Indels的最佳实践及Strelka等软件展开学习,了解NGS比对、比对后处理、germline和somatic calling的方法,并选择经典的数据集进行方法验证与讨论。
二、 变异检测研究-SV/CNV
SV通常定义为至少50个核苷酸碱基(50 bp) 以上的基因组变异,大致包括删除、插入、复制、倒位和易位。SV不仅在表现形式和检测方法上更为复杂,而其影响各不相同,部分SV与特定的肿瘤类型息息相关,而另一部分可能在许多肿瘤中存在。Somatic SV的数量、范围和类型能提供对肿瘤生成机制的见解。SV也可能成为治疗目标,或用作预后标记物。这里我们将介绍和学习肿瘤结构变异检测的方法。
三、 肿瘤免疫治疗的生物信息分析
肿瘤免疫治疗是目前最被寄予厚望的治疗方法,包括PD-1/PD-L、肿瘤疫苗等多种药物在临床研究中取得不错的结果。然而TMB的计算、新生抗原、免疫微环境的检测是其非常重要的环节。这里使用权威数据来开展讨论与实践,这些分析方法。
四、 肿瘤克隆性分析与肿瘤进化
肿瘤具有强烈时空异质性,其细胞可以形成具有遗传差异的克隆群。为了细致了解肿瘤,我们需要对癌症的不同时期(时间)或对不同位置(空间)进行取样测序,检测突变,根据突变的位置、频率和拷贝数信息进行聚类和进化分析。这里我们介绍相关的工具与方法。
五、 高频突变基因与驱动基因检测
高频突变基因与驱动基因的检测是研究肿瘤发生发展的一种重要的研究手段,相关的基因可能对于肿瘤有重要的驱动作用。这里我们来学习相关的软件与方法。
六、 肿瘤数据库学习与介绍
介绍一下肿瘤分析中必要的一些数据库,TCGA、靶药数据库等,了解必要的肿瘤数据库资源。
这个课程是高阶课程,需要有一些linux及编程基础才可以。如果您确实对这个课程感兴趣,但是没有编程及linux基础的话,我们可以提供我们自己录制的相关视频课程进行自学python及linux课程。对于无基础学员,通过我们的视频的学习与练习大约一周内就可以跟上课程要求了。
本课程适合从事医学肿瘤领域或者有志于往该方向发展,有一定Linux基础(能够进行基本的系统操作,并且可以使用命令行)并且在接下来的两个月内能够拿出足够多的时间复习及练习的同学。
那么这个课程学习后,我们觉得您应该有以下几方面的收获:
- 掌握肿瘤变异calling与注释的基本方法,对于影响变异的特殊因素及其解决方案有一定的认识。
- 掌握肿瘤免疫NGS相关分析,学会肿瘤免疫治疗的队列景观分析的研究方法。
- 掌握肿瘤克隆与进化的研究方法与手段,解析肿瘤队列挖掘关键基因,结合数据库进行解读以发表高水平研究的套路与方法。
- 适当拓展肿瘤NGS多组学研究的方法与手段。
最终可实现初步具备开展肿瘤变异研究的相关课题或者搭建与开发肿瘤相关应用产品的能力。