单细胞转录组技术毫无疑问是目前科研界的“当红炸子鸡”,但绝大部分人使用单细胞转录组的数据其实就当做是了流式细胞仪看不同细胞亚群组成的比例而已。这也就是为什么绝大部分单细胞转录组分析教程就是:数据质控,降维,聚类,分群,细胞亚群生物学注释。只要你的实验设计ok,这样的分析绰绰有余了,可以拿到一个落脚点去做实验验证一下或者公共数据库的验证。
但是流式细胞仪看不同细胞亚群组成的比例成本要低很多,而且仪器本身提供了集成式软件直接分析好数据,给出亚群比例,如下:
上图参考文章是:Scientific RepoRt (2020) 10:356 | https://doi.org/10.1038/s41598-019-57114-2
说到细胞组成比例,其实很多单细胞实验本身就结合了FACS筛选,因为绝大部分肿瘤相关单细胞研究的文章的第一次分群按照 :
- immune (CD45+,PTPRC),
- epithelial/cancer (EpCAM+,EPCAM),
- stromal (CD10+,MME,fibo or CD31+,PECAM1,endo)
但是如果咱们研究的落脚点并不是immune细胞,就可以使用CD45抗体来筛选富集细胞, 或者说我们关心的仅仅是肿瘤免疫微环境,就可以使用CD45抗体来阳选富集细胞,然后去单细胞转录组测序拿到数据后再分析。
最近看到一个文章是:Molecular Pathways of Colon Inflammation Induced by Cancer Immunotherapy. Cell 2020 Aug 6;182(3):655-671.e22. PMID: 32603654 就是如此,我注意到,它workflow写的是经过了CD3和CD45两次FACS筛选,如下所示 :
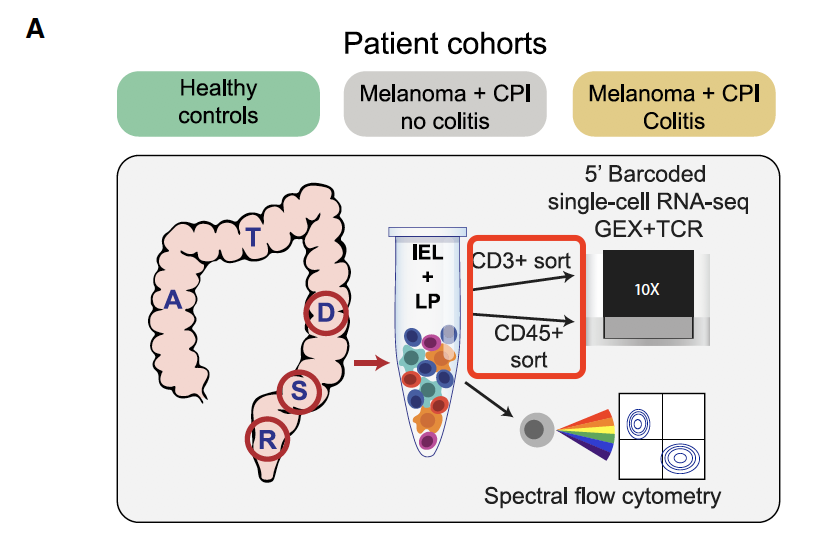
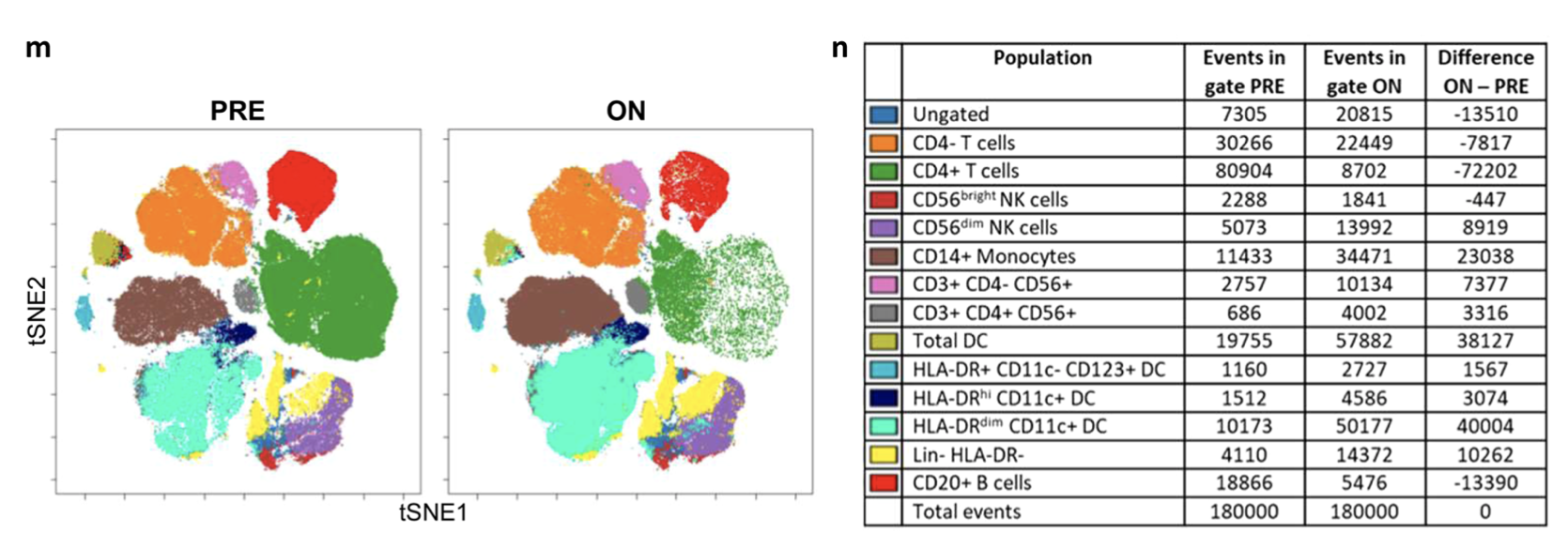
但是我看它分群的时候,居然还是有T细胞之外的各种免疫细胞,比如B细胞、masd,myeloid,等等。
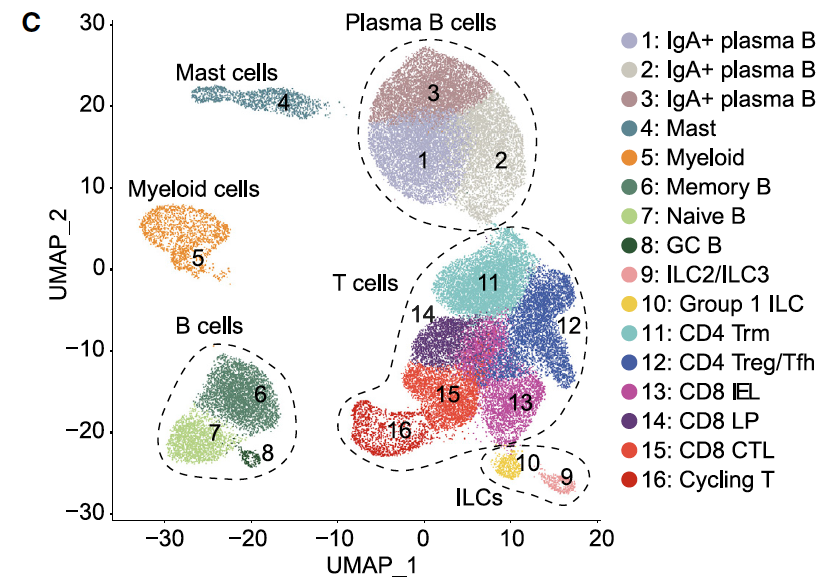
后来我仔细看,才明白,他们是做两个两次独立的FACS,独立的单细胞转录组数据。
首先是22个病人的CD3单次筛选后的单细胞RNA-seq, 22 patients from 3 different cohorts (Normal control, +CPI no colitis, +CPI colitis)
GSM4288826 +CPI colitis C1 sample CD3 RNA-seq
GSM4288827 +CPI colitis C2 sample CD3 RNA-seq
GSM4288828 +CPI colitis C3 sample CD3 RNA-seq
GSM4288829 +CPI colitis C4 sample CD3 RNA-seq
GSM4288830 +CPI colitis C5 sample CD3 RNA-seq
GSM4288831 +CPI colitis C6 sample CD3 RNA-seq
GSM4288832 +CPI colitis C7 sample CD3 RNA-seq
GSM4288833 +CPI colitis C8 sample CD3 RNA-seq
GSM4288834 +CPI no colitis NC1 sample CD3 RNA-seq
GSM4288835 +CPI no colitis NC2 sample CD3 RNA-seq
GSM4288836 +CPI no colitis NC3 sample CD3 RNA-seq
GSM4288837 +CPI no colitis NC4 sample CD3 RNA-seq
GSM4288838 +CPI no colitis NC5 sample CD3 RNA-seq
GSM4288839 +CPI no colitis NC6 sample CD3 RNA-seq
GSM4288840 Normal control CT1 sample CD3 RNA-seq
GSM4288841 Normal control CT2 sample CD3 RNA-seq
GSM4288842 Normal control CT3 sample CD3 RNA-seq
GSM4288843 Normal control CT4 sample CD3 RNA-seq
GSM4288844 Normal control CT5 sample CD3 RNA-seq
GSM4288845 Normal control CT6 sample CD3 RNA-seq
GSM4288846 Normal control CT7 sample CD3 RNA-seq
GSM4288847 Normal control CT8 sample CD3 RNA-seq
然后是 18 个病人的 CD45单次筛选后阳性的单细胞RNA-seq :
GSM4288848 +CPI colitis C1 sample CD45 RNA-seq
GSM4288849 +CPI colitis C2 sample CD45 RNA-seq
GSM4288850 +CPI colitis C3 sample CD45 RNA-seq
GSM4288851 +CPI colitis C4 sample CD45 RNA-seq
GSM4288852 +CPI colitis C5 sample CD45 RNA-seq
GSM4288853 +CPI colitis C6 sample CD45 RNA-seq
GSM4288854 +CPI no colitis NC1 sample CD45 RNA-seq
GSM4288855 +CPI no colitis NC2 sample CD45 RNA-seq
GSM4288856 +CPI no colitis NC3 sample CD45 RNA-seq
GSM4288857 +CPI no colitis NC4 sample CD45 RNA-seq
GSM4288858 +CPI no colitis NC6 sample CD45 RNA-seq
GSM4288859 Normal control CT1 sample CD45 RNA-seq
GSM4288860 Normal control CT2 sample CD45 RNA-seq
GSM4288861 Normal control CT3 sample CD45 RNA-seq
GSM4288862 Normal control CT4 sample CD45 RNA-seq
GSM4288863 Normal control CT6 sample CD45 RNA-seq
很容易理解,CD45单次筛选后阳性的单细胞RNA-seq,是几乎全部的免疫细胞啦,所以有各种各样的B细胞、masd,myeloid,等都是可以理解的。
但是呢,CD3单次筛选后的单细胞RNA-seq,理论上只有T细胞,不可能分群出来其它细胞类型!
现在问题来了,两次不同的筛选策略,拿到的单细胞转录组数据,分群后一些细胞的比例会发生变化吗?
历年学徒作业目录如下:
- 生信编程直播课程优秀学员作业展示1
- 生信编程直播课程优秀学员学习心得及作业展示3
- 生信编程直播课程优秀学员作业展示2
- 给学徒的GEO作业
- 这个WGCNA作业终于有学徒完成了!
- 上次说的gmt函数(学徒作业)
- 拖后腿学徒居然也完成作业,理解RNA-seq数据分析结果
- 肿瘤外显子视频课程小作业
- ChIPseq视频课程小作业
- Agilent芯片表达矩阵处理(学徒作业)
- 学徒作业:TCGA数据库单基因gsea之COAD-READ
- 学徒作业-在CCLE数据库里面根据指定基因在指定细胞系里面提取表达矩阵
- 学徒作业-指定基因在指定组织里面的表达量热图
- 学徒作业-我想看为什么这几个基因的表达量相关性非常高
- 学徒作业:给你8个甲基化探针, 你在tcga数据库进行任意探索
- 学徒作业-根据我的甲基化视频教程来完成2015-NPC-methy-GSE52068研究
- RNA芯片和测序技术的比较(学徒作业)
- 学徒作业-单基因的tcga数据挖掘分析
- ATCC终于出来了organoids资源
- 拿到7个DDR通路的基因集-学徒作业
- 绘图本身很简单但是获取数据很难
- 都说lncRNA只有部分具有polyA尾结构,请证明
- 学徒作业-hisat2+stringtie+ballgown流程
- 学徒任务-探索DNA甲基化的组织特异性
- 用WES和RNA-Seq数据提取到的somatic SNVs不一致
- 《GEO数据挖掘课程》配套练习题