前面的教程里面,我们首先根据 CNS图表复现08—肿瘤单细胞数据第一次分群通用规则进行了初步分群,如下所示:
immune (CD45+,PTPRC), epithelial/cancer (EpCAM+,EPCAM), and stromal (CD10+,MME,fibo or CD31+,PECAM1,endo)
然后根据CNS图表复现06-根据CellMarker网站进行人工校验免疫细胞亚群 进行了免疫细胞细分亚群,但是我注意到,其实文章给定了一下他们自己的收集整理好的标记基因作为他们文章的分群依据,如下:
Table Description
1. General Cell Markers , General markers used for differing between non-immune and immune cell types as well as non immuen epithelial cell types
2. COSMIC mutation list , COSMIC Tier 1 genes and overlap with genes used in clincal DNA assays
3. Cancer Cell Signature Genes ,Gene lists of each cancer signature
4. Immune Markers Markers , used for differing between primary immune cell types
现在我们就校验一下原文的细胞亚群的标记基因的可靠性:
首先看 General Cell Markers
首先从 General Cell Markers , General markers used for differing between non-immune and immune cell types as well as non immuen epithelial cell types拿到基因名字:
代码如下:
rm(list=ls())
options(stringsAsFactors = F)
library(Seurat)
library(ggplot2)
load(file = 'first_sce.Rdata')
load(file = 'phe-of-first-anno.Rdata')
sce=sce.first
table(phe$immune_annotation)
sce@meta.data=phe
sce@meta.data$new=paste(phe$immune_annotation,phe$seurat_clusters)
genes_to_check=c('PTPRC','CD3G','CD3E','CD79A','BLNK','CD68','CSF1R','MARCO','CD207','PMEL','MLANA','PECAM1','CD34','VWF','EPCAM','SFN','KRT19','ACTA2','MCAM','MYLK','MYL9','FAP','THY1','ALB')
p3 <- DotPlot(sce, features = genes_to_check,
assay='RNA',group.by = 'new' ) #+ coord_flip()
p3
可以很清楚的看到,高表达ALB基因的Hepatocytes被我划分到了stromal细胞大群,是需要区分出来的。而且高表达PMEL和MLANA的Melanocytes也被我划分到了stromal细胞大群,是需要区分出来的。
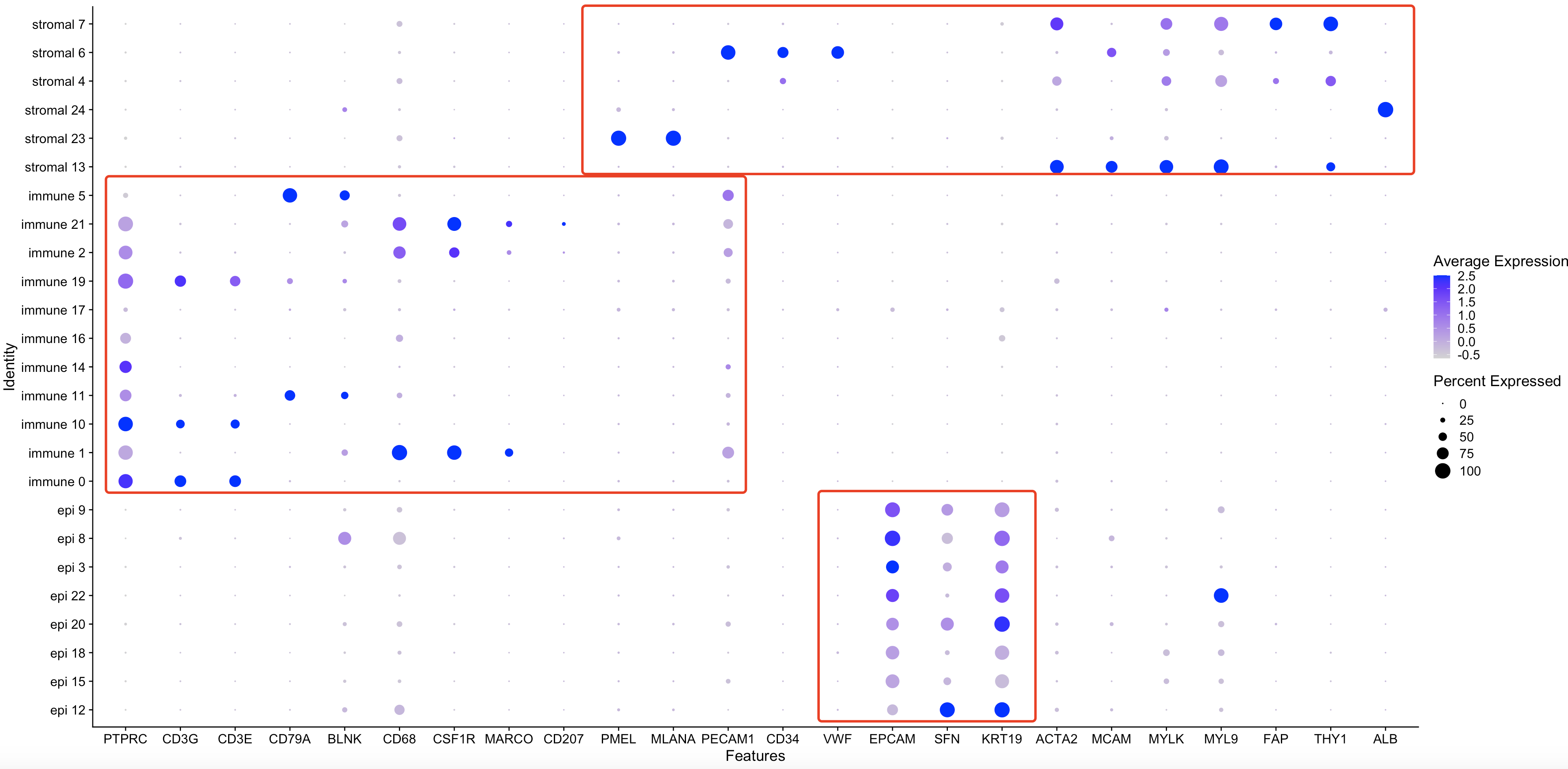
而且有一群细胞,既表达EPCAM等上皮细胞的标记基因,也表达MYL9这个Fibroblasts的基因,很有可能是并不纯粹的细胞亚群,或者说是双细胞情况。
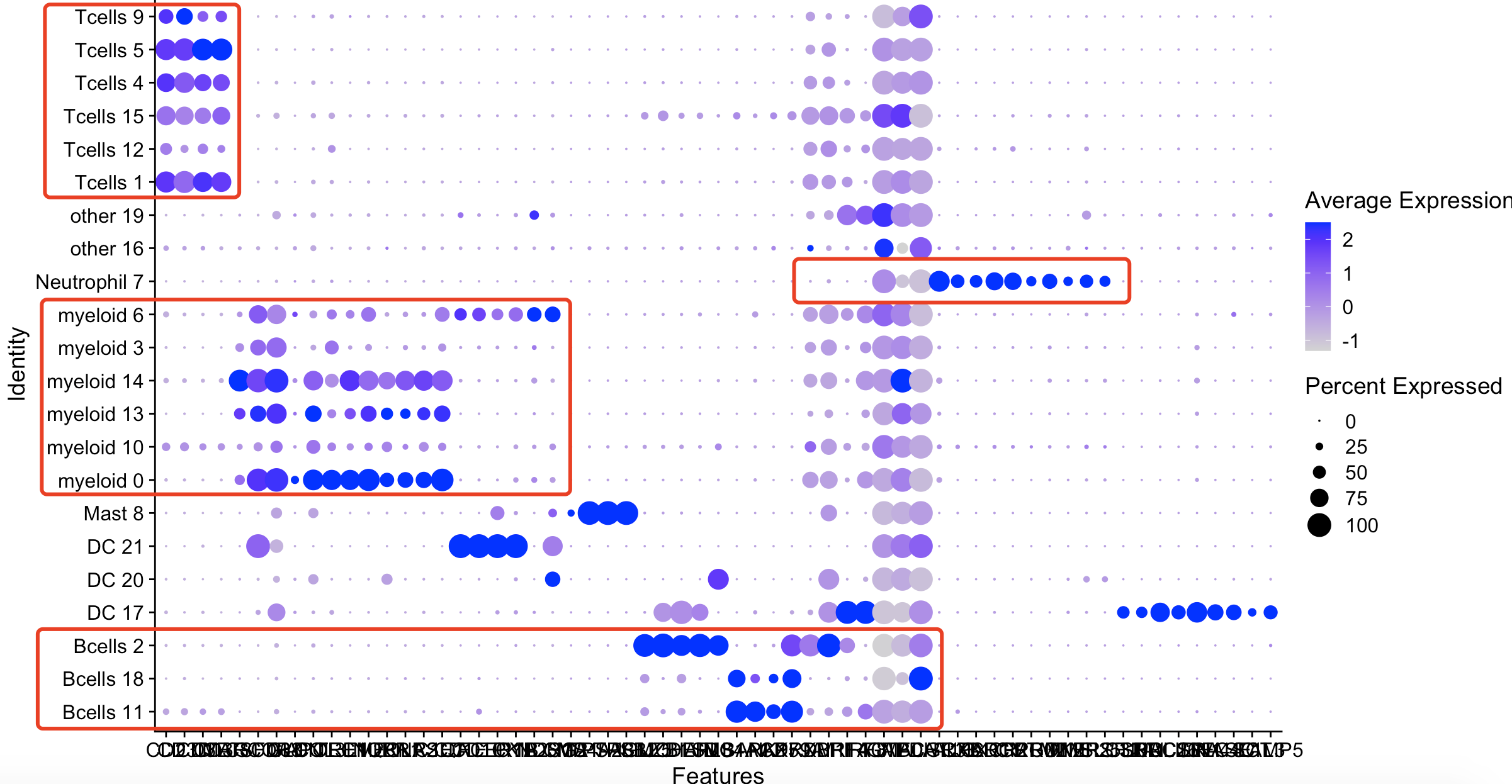
然后看 Immune Markers Markers
首先从 Immune Markers Markers , used for differing between primary immune cell types 拿到基因名字。
承接上面的代码,如下:
cells.use <- row.names(sce@meta.data)[which(phe$immune_annotation=='immune')]
length(cells.use)
sce <-subset(sce, cells=cells.use)
sce
load(file = 'phe-of-subtypes-Immune-by-manual.Rdata')
sce@meta.data=phe
table(phe$immuSub)
table(phe$immuSub,phe$seurat_clusters)
sce@meta.data$new=paste(phe$immuSub,phe$seurat_clusters)
table(sce@meta.data$new)
genes_to_check=c( 'CD2','CD3D','CD3E','CD3G','MARCO','CSF1R','CD68','GLDN','APOE','CCL3L1',
'TREM2','C1QB','NUPR1','FOLR2','RNASE1','C1QA','CD1E','CD1C','FCER1A','PKIB',
'CYP2S1','NDRG2','CMA1','MS4A2','TPSAB1','TPSB2','IGLL5','MZB1','JCHAIN','DERL3',
'SDC1','MS4A1','BANK1','PAX5','CD79A','PRDM1','XBP1','IRF4','MS4A1','IRF8','ACTB',
'GAPDH','MALAT1','FCGR3B','ALPL','CXCR1','CXCR2','ADGRG3','CMTM2','PROK2','MME','MMP25',
'TNFRSF10C','SLC32A1','SHD','LRRC26','PACSIN1','LILRA4','CLEC4C','DNASE1L3',
'CLEC4C','LRRC26','SCT','LAMP5')
genes_to_check=unique(genes_to_check)
p4 <- DotPlot(sce, features = genes_to_check,
assay='RNA',group.by = 'new' ) #+ coord_flip()
p4
在所有的细胞亚群,都表达的基因是3个Housekeeping,分别是: ‘ACTB’, ‘GAPDH’,’MALAT1’