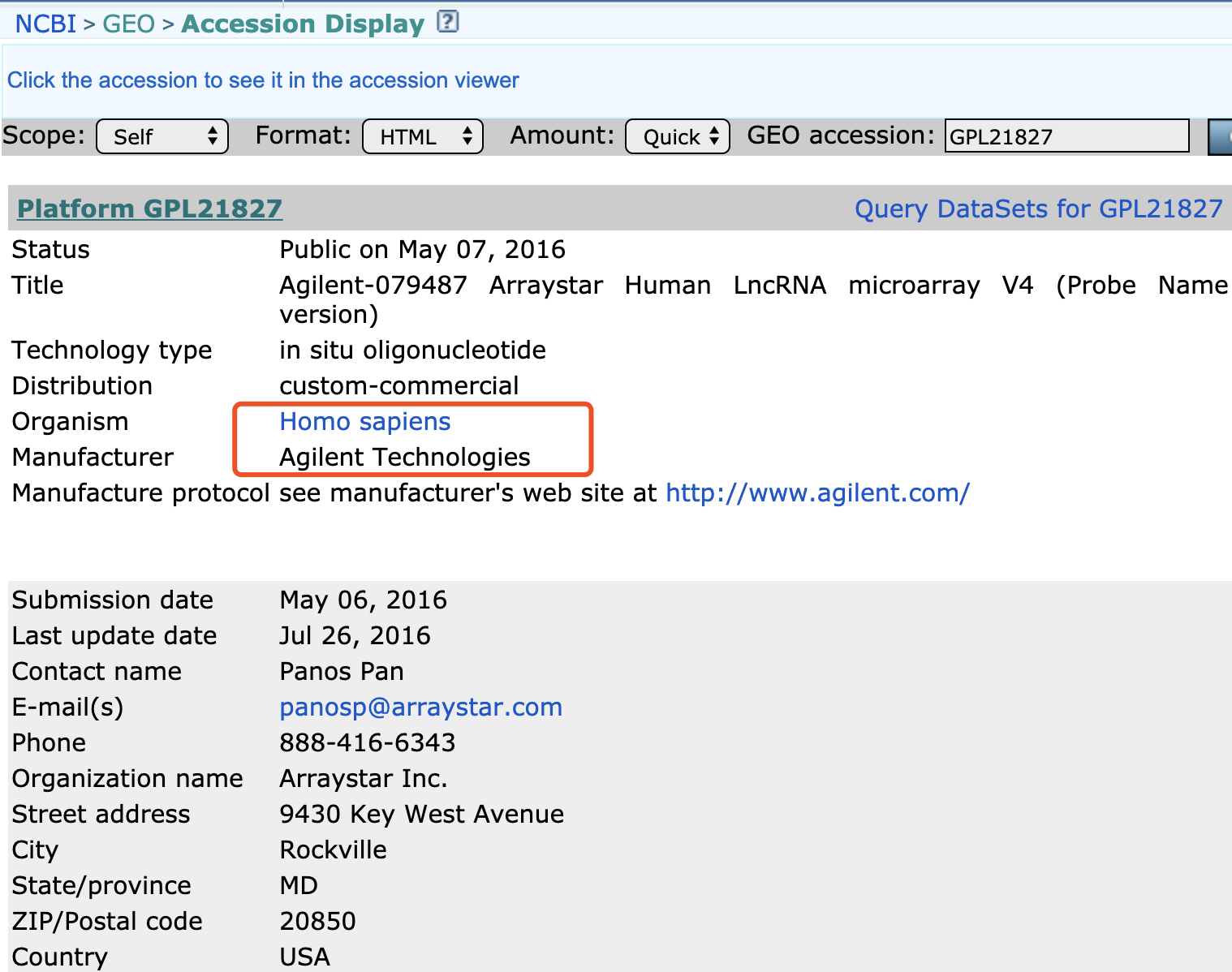
最近在对GEO数据库的全部GPL平台的芯片探针序列进行批量重新注释的时候,发现如果工具芯片自带的物种信息来自动化选择参考基因组,居然还会出现某个芯片探针比对率非常低的情况, 比如GPL21827这个平台:
60898 reads; of these:
60898 (100.00%) were unpaired; of these:
59099 (97.05%) aligned 0 times
1753 (2.88%) aligned exactly 1 time
46 (0.08%) aligned >1 times
2.95% overall alignment rate
因为在GEO数据库,它居然被记录为mouse这个物种,但是它明明是human啊!
Agilent-079487 Arraystar Human LncRNA microarray V4 (Probe Name version)
GPL21827
Public on May 07 2016
2016/5/6
2016/5/7
in situ oligonucleotide
Mus musculus
Agilent Technologies
实在是太诡异了:https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL21827
可是我在GEO官网查询它: Agilent-079487 Arraystar Human LncRNA microarray V4 (Probe Name version)
物种又是human。
这并不是唯一的比对率低的情况:
一款circRNA芯片: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL23467
170351 reads; of these:
170351 (100.00%) were unpaired; of these:
169391 (99.44%) aligned 0 times
811 (0.48%) aligned exactly 1 time
149 (0.09%) aligned >1 times
0.56% overall alignment rate
一款miRNA芯片 : https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GPL8180
380 reads; of these:
380 (100.00%) were unpaired; of these:
215 (56.58%) aligned 0 times
138 (36.32%) aligned exactly 1 time
27 (7.11%) aligned >1 times
43.42% overall alignment rate