发表于2017年,杂志是 JOURNAL OF CLINICAL ONCOLOGY 影响因子26.303 , 文章是 CpG Methylation Signature Predicts Recurrence in Early-Stage Hepatocellular Carcinoma: Results From a Multicenter Study 亮点应该是自己的数据,然后使用了两个机器学习算法:
- LASSO, Least Absolute Shrinkage and Selector Operation;
- SVM-RFE, Support Vector Machine-Recursive Feature Elimination;
前面我们讲解了一篇2013年多组学数据探索乳腺癌细胞系药物敏感性使用的也是两个机器学习算法,不过是LS-SVM和RF,但是也有借鉴意义。课题设计
自己的450K甲基化芯片数据上传到了:GSE75041
本项目共纳入 576 patients with Early-stage hepatocellular carcinoma (E-HCC) ,其中 - 66 tumor samples were analyzed using the Illumina Methylation 450k Beadchip.
- internal cohort (n = 141) and two external cohorts (n = 191 and n =104).
也就是先小队列做450K拿到感兴趣的甲基化位点,然后扩大队列只测量感兴趣的甲基化位点证明自己拿到的位点是有临床价值的,整体课题设计如下:
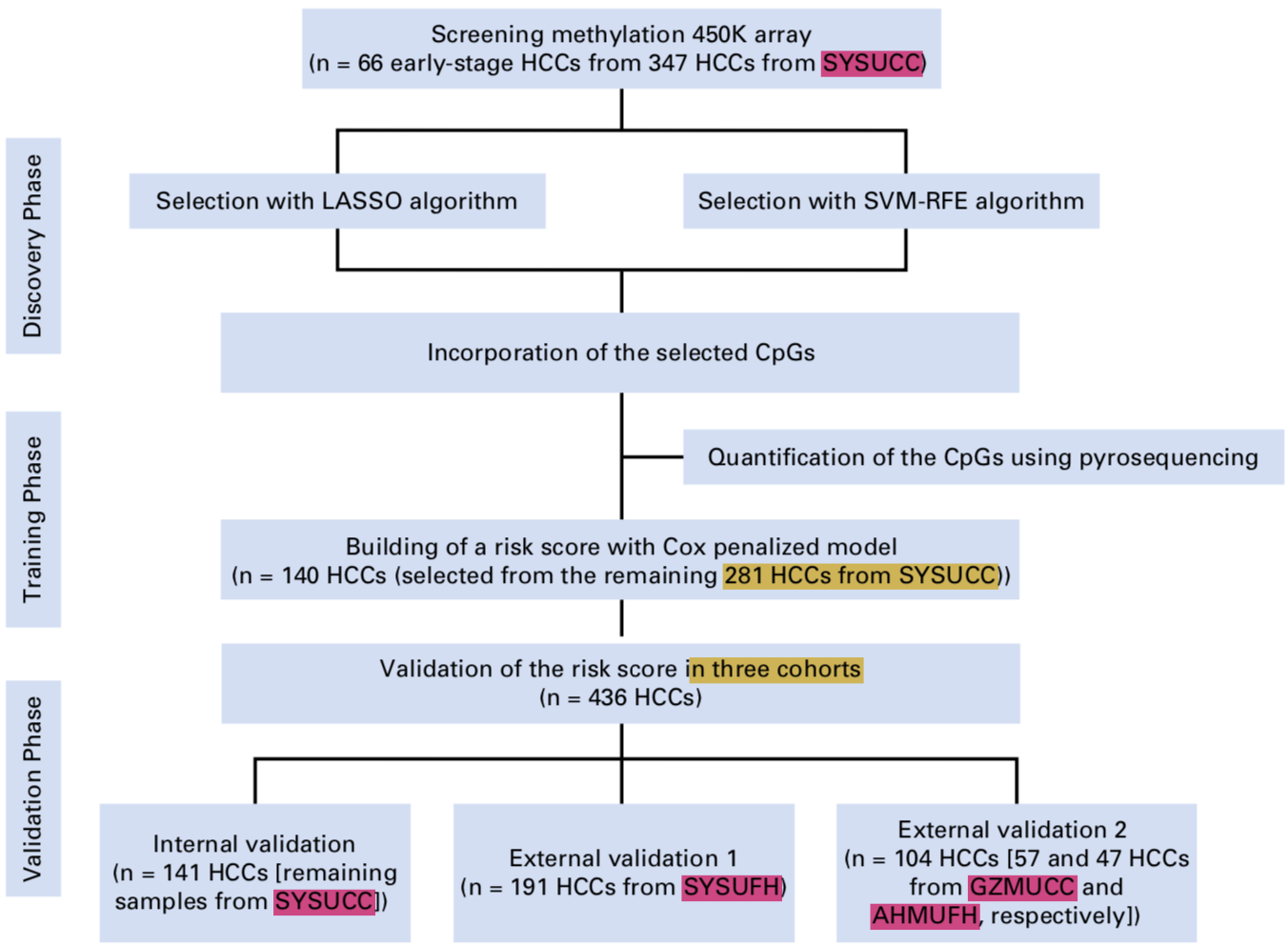
项目纳入的病人来源: - 347 E-HCC samples at the Sun Yat-sen University Cancer Center (SYSUCC)
- 295 samples at three independent centers as follows:
- 191 samples from the First Affiliated Hospital of Sun Yat-sen University
- 57 samples from Guangzhou Medical University Cancer Center (GZMUCC)
- 47 samples from the First Affiliated Hospital of Anhui Medical University (AHMUFH).
文章的introduction部分肯定是介绍 E-HCC疾病的重要性,还有甲基化信号的重要性。
当然,也不落俗套的在 The Cancer Genome Atlas (TCGA) database 数据库进行验证。数据处理
首先,复发与否的66个肿瘤样本数据找差异甲基化位点,得到 a list of 2,550 differential CpGs
然后使用 LASSO algorithm to identify a set of 30 CpGs
接着使用 SVM-RFE algorithm and selected a set of 30 CpGs
两个算法有14个CpG位点的交集,如下图所示:
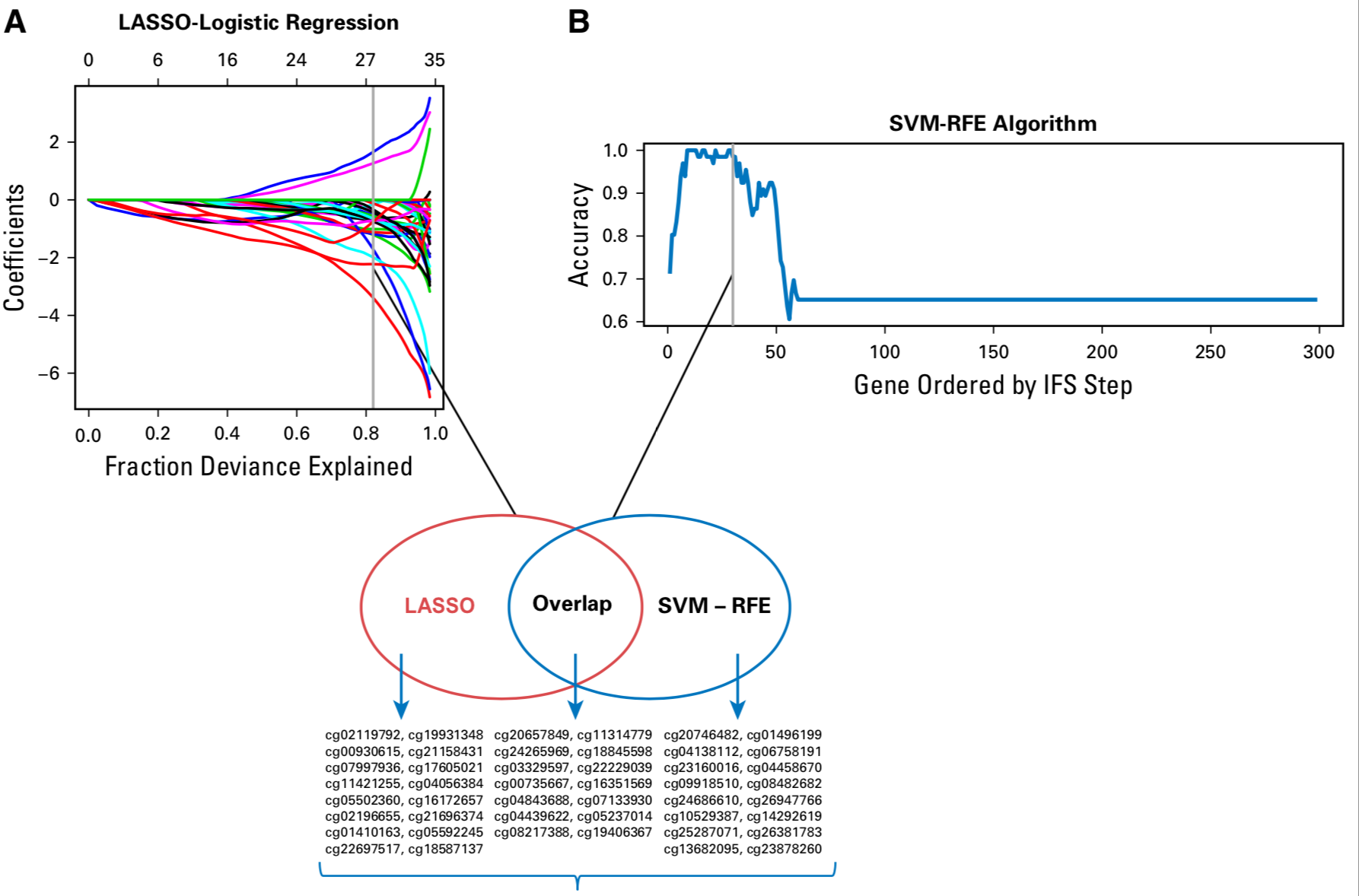
其中并集是46个,可以看热图如下:
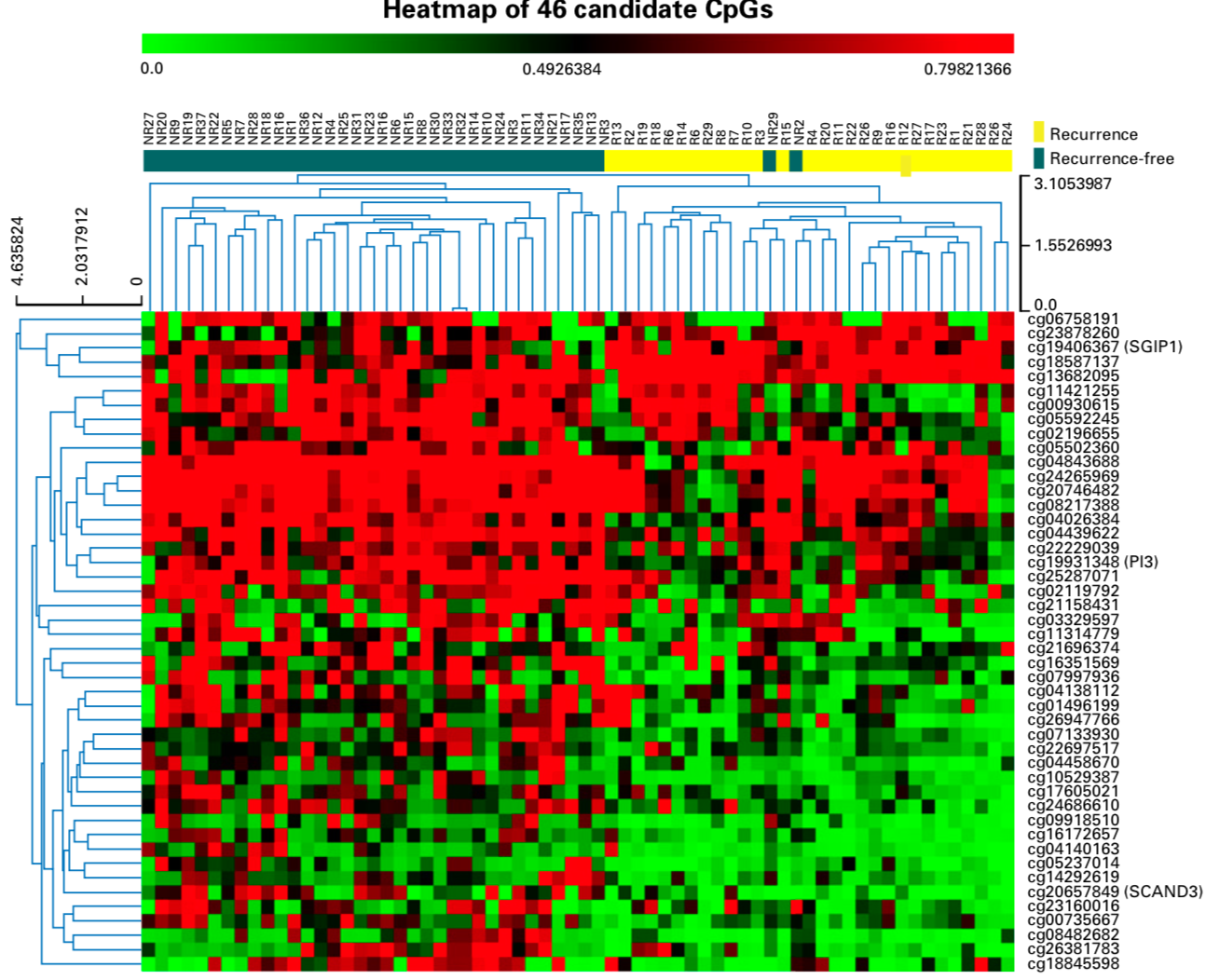
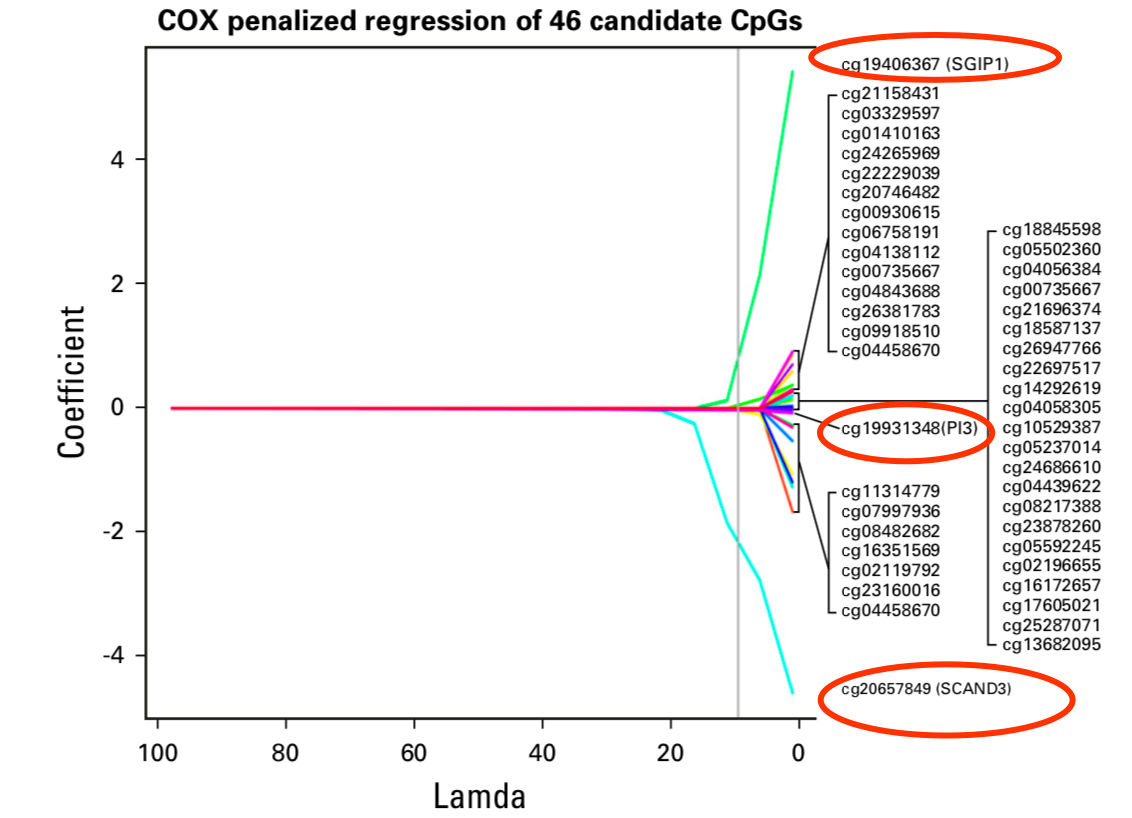
继续使用 penalized Cox regression model ,最后缩小到3个甲基化位点:
- cg20657849, SCAN domain containing 3 (SCAND3)
- cg19406367, Src homology 3-domain growth factor receptor-bound 2-like interacting protein 1 (SGIP1)
- cg19931348 ,peptidase inhibitor 3 (PI3)
算法的效果如下;
同时也根据这3个甲基化位点,构建了风险模型公式:risk score = (0.104 × methylation level of SGIP1) + (−1.125 × methylation level of SCAND3) + (−0.085 × methylation level of PI3).
并且称之为: a methylation-based signature for patients with E-HCC (MSEH)
然后就可以去验证集里面去看看预测效果。生存分析验证模型效果
在开头我们介绍的数据集里面,作者都使用了生存分析,很显著的发现这3个甲基化位点组成的a methylation-based signature for patients with E-HCC (MSEH) 具有很好的区分效果,如下图:

因为作者验证的数据集已经有3个了,所以在TCGA的验证作者只是放在附件。In addition, the predictive value of MSEH was validated further in the TCGA data. MSEH successfully discriminated 125 patients with TNM stage I into high-risk and low-risk groups in terms of both RFS and OS (P , .001, P = .043, respectively; Data Supplement).
感兴趣的朋友也可以很容易去下载TCGA的肝癌的甲基化信号矩阵,来根据这3个甲基化位点组成的a methylation-based signature for patients with E-HCC (MSEH) 来进行验证。