发表于2018年的NC,文章是:Multi-omics profiling of younger Asian breast cancers reveals distinctive molecular signatures 文章特色就是测了 187 primary tumors from a Korean BC cohort (SMC) 的转录组和外显子组数据。数据上传在:EGAS0000100262 和 GSE113184 .
背景知识
了解TCGA计划中的乳腺癌研究现状
了解一个事实:西方人的乳腺癌患者中15−30% 是绝经期前,而亚洲人这一比例接近50%,人种差异需要解释,而且影响治疗策略的选择,因为Breast cancers arising in younger patients (YBC)通常更恶性。特别的,ER阳性的YBC趋向于抵抗内分泌治疗。但是为什么YBC患者生存更糟糕却研究不够。
得益于多组学技术的发展以及TCGA计划,西方人群体的乳腺癌的分子分型和遗传特性研究的比较清楚。也有一些研究比较了年轻一点的乳腺癌患者和年长一点的乳腺癌患者。
- ESR1基因的转录表达水平和蛋白表达水平都是YBC大于OBC,而且YBC趋向于超甲基化。
- YBC群体的GATA3突变多,而OBC群体里面 CDH1居多。
- ER阳性的YBC表现出integrin/laminin, EGFR signaling, and TGF-β通路激活,还有proliferation, stem cell, and endocrine resistance通路也是在YBC过于激活。
但是关于亚洲人的研究一直比较少,或者涉及的人群样本量小: - 其中一个研究纳入 113 Middle Eastern women and identified 63 genes specific to tumors in young women
- 另外一个研究比较了 Chinese and Italian 乳腺癌的 基因表达和miRNA表达。
缺乏多组学数据,而且也缺乏亚洲人的年轻乳腺癌患者数据,所以作者做了这个研究。两个cohort的病人年龄及亚型区别
TCGA计划里面有1116个美国乳腺癌患者,而作者是SMC里面有186个患者,都是WES数据加上RNA-seq数据。
首先TCGA按照年龄分3类 - YBC (age ≤ 40, n = 181)
- IBC (40 < age ≤ 60, n = 562)
- OBC (age > 60, n = 535).
然后韩国人的研究分2类 - YBC (age ≤ 40, n = 125)
- IBC (age > 40, n = 62)
可以看到区别很明显,如下;
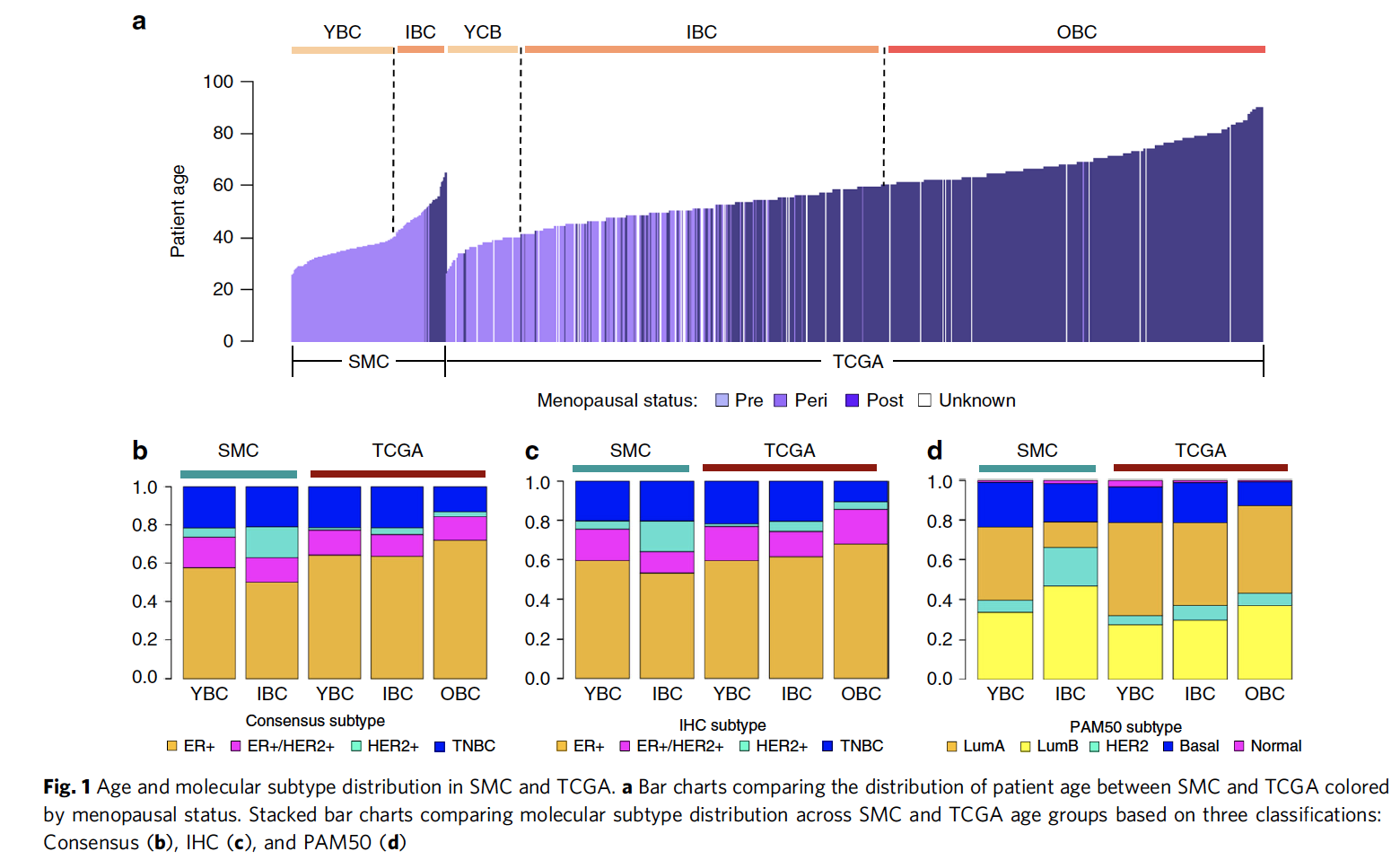
分子分型的三种方式:
分别是:
- ER and HER2 immunohistochemistry (IHC) analyses
- gene expression classifier (PAM50)
- a naive molecular classifier (NMC) based on ESR1, PGR, ERBB2 gene expression and ERBB2 copy number data.
这3种亚型在SMC和TCGA数据集的比例区别也如上图所示。TCGA公共数据的临床三线表
有现成的R包可以做,直接在TCGA官网下载临床信息,筛选后即可直接生成表格,作者他们自己的SMC数据集也可以做临床三线表,这个是所有人群队列研究必备的,如下:
胚系遗传性的致病突变
BRCA1/BRCA2 是乳腺癌患者最出名的易感基因,之前的数据是西方高加索人群乳腺癌群体携带率是1~5%,而亚洲人高达3−7% ,所以作者也检查了自己的韩国乳腺癌队列的胚系遗传性的致病突变情况,主要是寻找:
- truncate protein reading frame or reported as a disease-causing variant in ClinVar
- 13 genes known to increase breast cancer susceptibility with high to moderate penetrance
发现携带率是 18.8% (35/186) ,其中BRCA1/2就明显在SMC里面多余在TCGA队列,10.8% of SMC but only 4.7% of TCGAsomatic突变
数据分析得到总共是6885个影响蛋白的突变,涉及到4949个基因,TMB平均是0.6,很明显 TCGA (1.4 ± 4.5) 是高于 SMC (0.90 ± 0.97) 的,因为TMB跟年龄相关,而且SMC队列以年轻人为主。
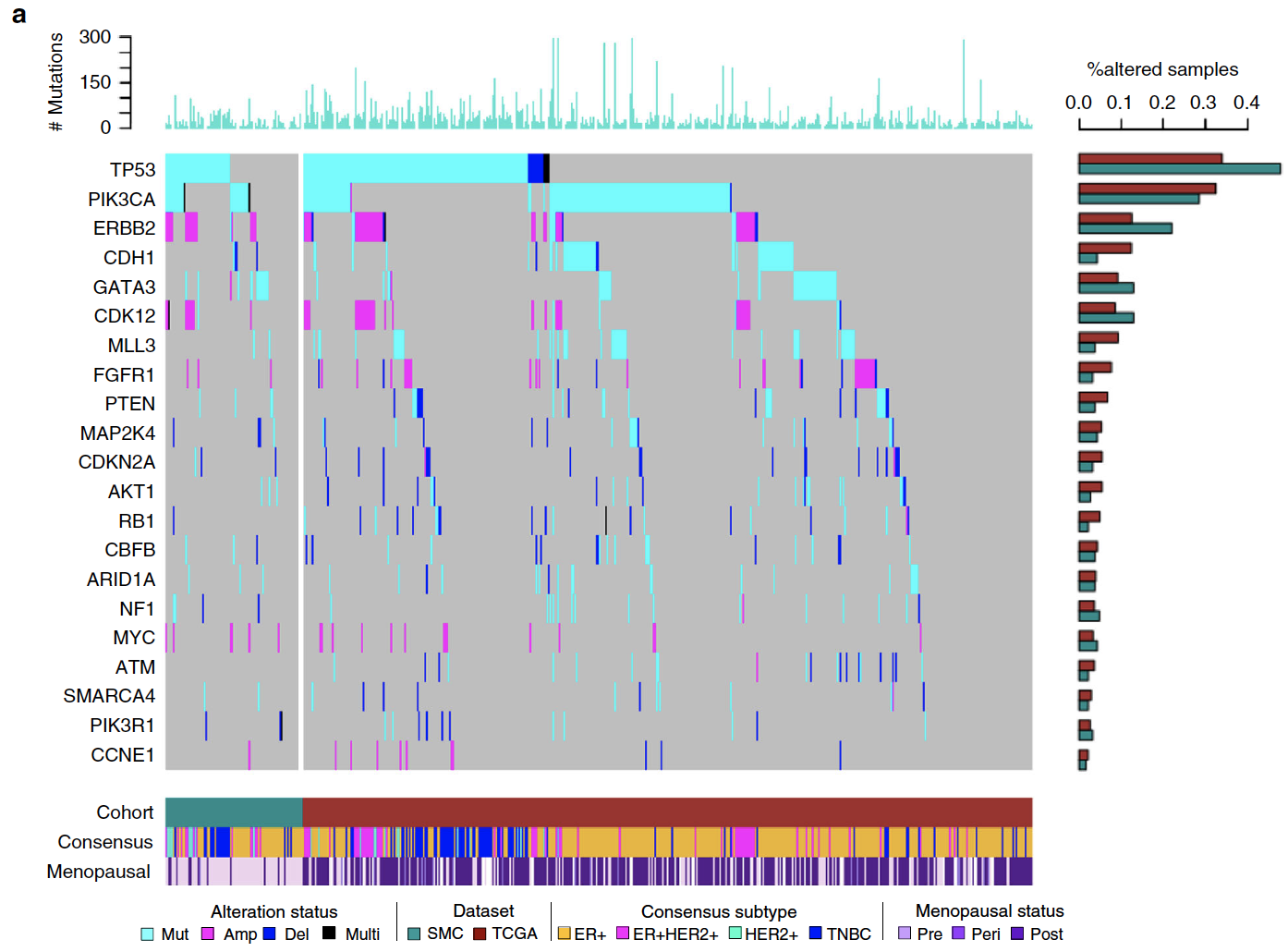
队列数量足够大,所以可以用MutSigCV 分析显著突变基因,得到6个,分别是:TP53, PIK3CA, GATA3, CBFB, PTEN, and CDH1 然后就一个个基因说明它在TCGA和SMC队列的区别。
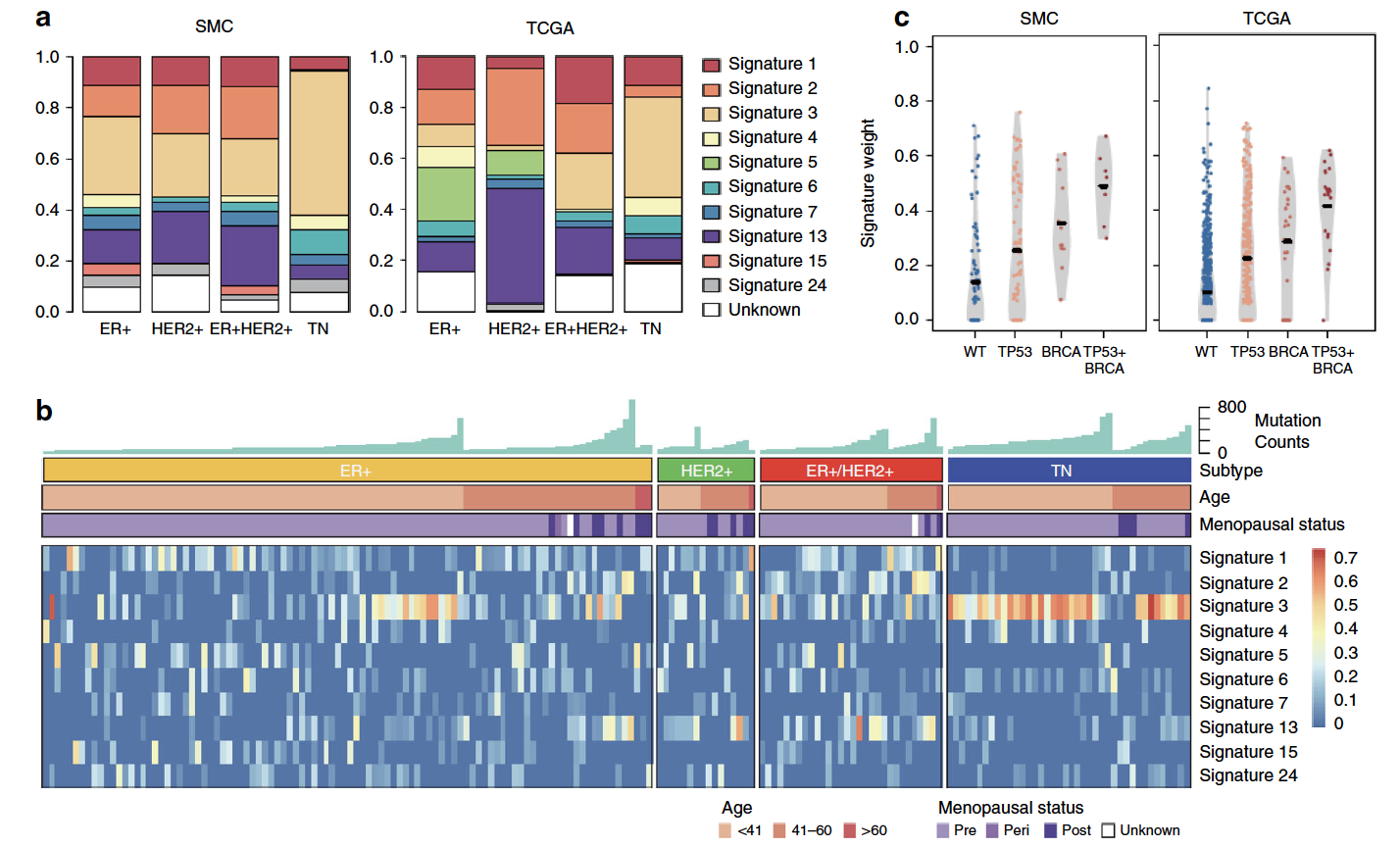
而且也可以比较两个队列的突变全景图。突变的signature
这个也是癌症研究的一个常见要点了,可以是2012的cosmic数据库的30个signatures作为库,使用成熟的R包来分析自己的数据集的突变情况,再跟TCGA的对比。
NMF模拟显微切割
首先纳入了 4 个数据集的共1678个样本,如下图:
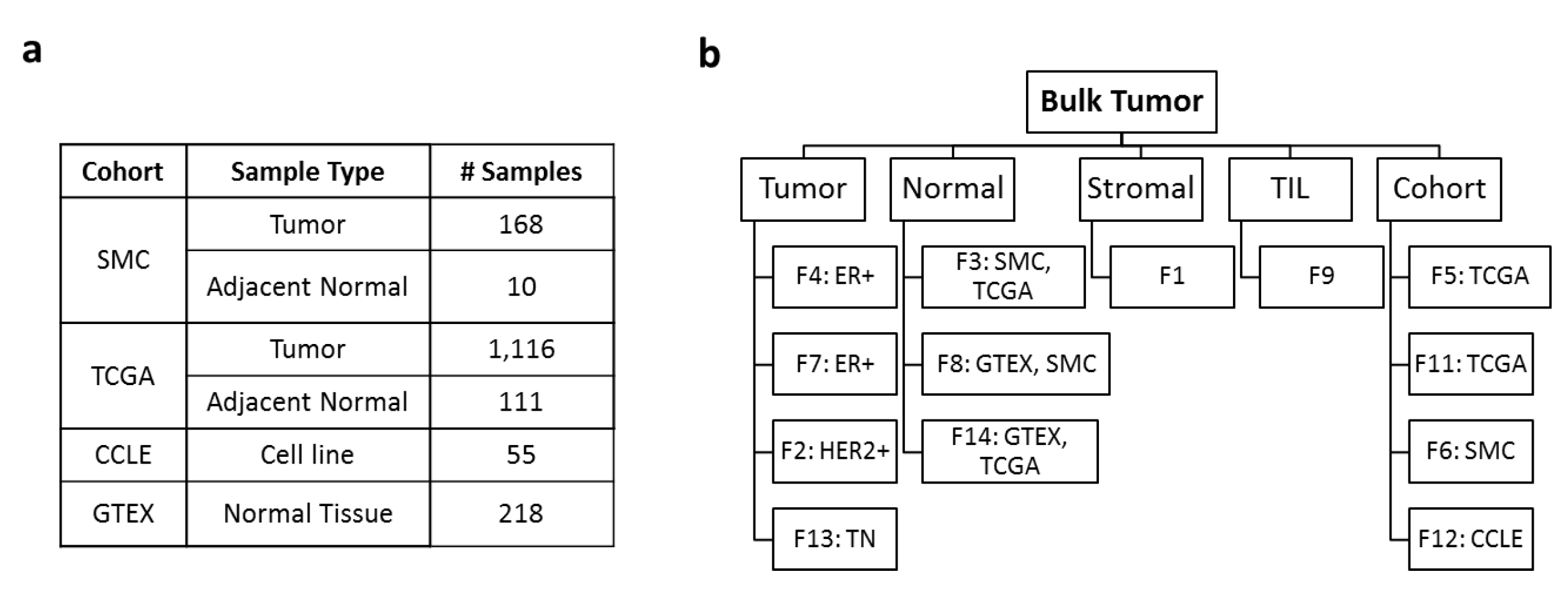
通过技术可以得到 13 NMF factors were attributed to 4 tissue compartments - tumor intrinsic
- stroma
- tumor infiltrating leukocyte (TIL)
- normal tissue.
其中有每个数据集都有一个自己独特的NMF因子,可以理解为批次效应,下面是不同的因子在不同的数据集的表现。
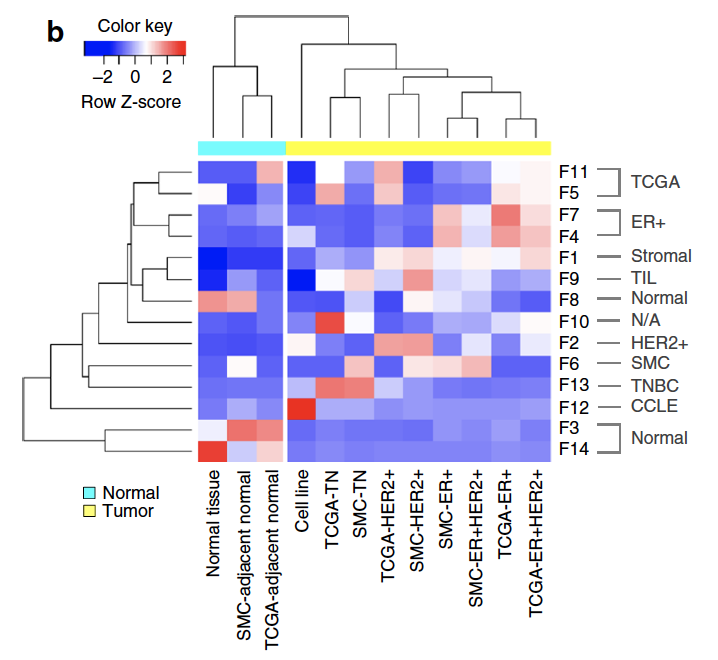
这个算法也被打包到R里面了,很容易复现这个数据分析流程。
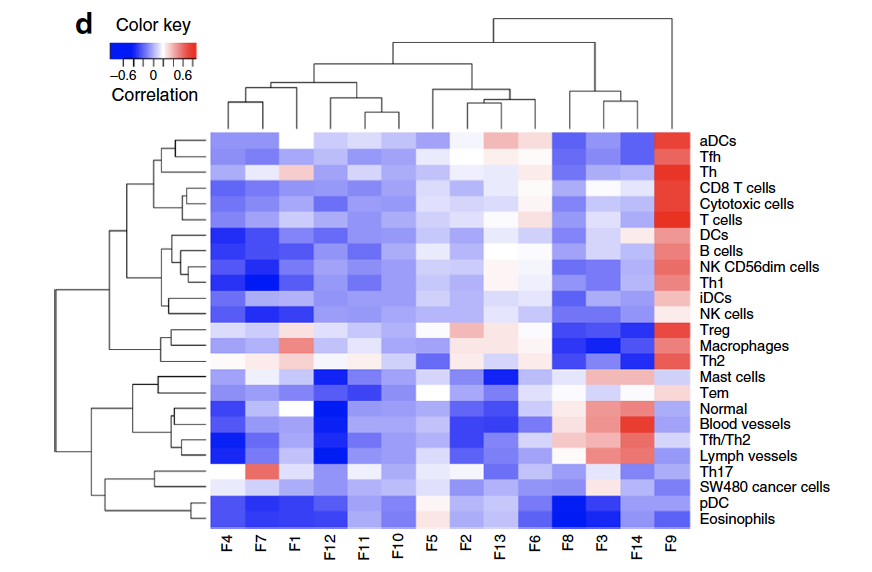
也可以把Bindea immune expression signature scores (GSVA) and NMF factors做一下相关性,这样可以解释NMF算法得到的这些因子的生物学意义。
差异分析及功能注释
作者这里采用了voom来对表达矩阵进行归一化成logCPM形式,然后使用limma做差异分析吗,定义FDR < 0.01 and |log2FC| > 0.2的基因为显著差异。
值得注意的是作者过滤掉了25%的低表达量基因以及25%的低SD基因。
使用GSVA算法,计算了MsigDB v5.1数据库的6475 gene sets - H (hallmark gene sets)
- C2 (curated gene sets)
- C5 (GO gene sets)
- C6 (oncogenic gene sets)
只有表达矩阵是比较方便下载的。附件信息比较丰富
虽然无法下载全外显子测序数据,但是作者团队的分析结果都在附件给出了。
Supplementary Data 1: SMC and TCGA sample annotation.
Supplementary Data 2: Clinical data summary of SMC and TCGA.
Supplementary Data 3: Molecular subtype distribution in SMC and TCGA. This table contains proportions of molecular subtypes (Consensus, IHC and PAM50) in different age groups and cohorts. Also included are statistical significances of differential distribution between different sample groups.
Supplementary Data 4: BRCA1/BRCA2 germline pathogenic mutations.
Supplementary Data 5: Mutation prevalence of cancer driver genes in SMC and TCGA. This table contains mutation frequencies (% samples) in different cohorts and age groups for significantly mutated genes. Also included are statistical significances of differential distribution and frequencies of germline pathogenic mutations for BRCA1 and BRCA2.
Supplementary Data 6: Somatic mutation predictions in SMC. Detailed annotations for 6,885 somaticprotein-altering mutations.
Supplementary Data 7: Significantly mutated genes. Significantly mutated genes identified by MutSigCVbased on combined mutations from SMC and TCGA (“Combined”), SMC mutations alone (“SMC”) and TCGAmutations alone (“TCGA”). n_nonsilent: number of protein-altering mutations. n_silent: number of silentmutations.
Supplementary Data 8: Somatic alteration prevalence of cancer driver genes in SMC and TCGA. This table contains the prevalence (% samples) of somatic alterations, including protein-altering substitutions,insertions, deletions, copy number amplifications and deletions, for frequently altered genes in different agegroups and cohorts. Amplification is defined as absolute copy number ≥ 6 and deletion as copy number ≤ 1.Also included are statistical significances of different group comparisons. Both direct comparisons with Fisher’s exact test and comparisons adjusting for tumor stage with logistic regression were performed. P-value was calculated using the Fisher’s exact test and FDR corrected using the Benjamini-Hochberg method.
Supplementary Data 9: Somatic alteration prevalence of oncogenic pathways. This table containsalteration prevalence of five BC related oncogenic pathways in different age groups and cohorts. A pathwaywas altered in a sample if one or more genes in that pathway harbor alteration in the sample. Pairwisecomparison of alteration prevalence was performed between different groups using the Fisher’s exact test.
Supplementary Data 10: Somatic mutation signatures. (a) Mutation signature distribution acrosssubtypes and cohorts. (b) Mutation signature correlation with mutation burden, number of protein-alteringsomatic mutations in each sample.
Supplementary Data 11: Pathway enrichment of NMF factors. Statistical significance for pairwiseassociations between DE pathways and NMF factors.
Supplementary Data 12: Differentially expressed genes and pathways. (a) Differentially expressed genes(a) and pathways (b) identified from two comparisons - SMC pre-menopausal vs. TCGA post-menopausal andSMC pre-menopausal vs. TCGA pre-menopausal.