来自于Florian的博客: https://scientificbsides.wordpress.com/
目录如下:
- Inferring tumour evolution 3 – Methods for single samples | Scientific B-sides
- Inferring tumour evolution 4 – methods for multiple samples of the same tumour | Scientific B-sides
- Inferring tumour evolution 5 – single cell data | Scientific B-sides
- Inferring tumour evolution 6 – What do we talk about when we talk about a clone? | Scientific B-sides
- Inferring tumour evolution 1 – The intra-tumour phylogeny problem
- Inferring tumour evolution 7 – the roots of metastasis
“Cancer evolves dynamically as clonal expansions supersede one another driven by shifting selective pressures, mutational processes, and disrupted cancer genes. These processes mark the genome, such that a cancer’s life history is encrypted in the somatic mutations present,”
有一个中文博客进行了简单翻译:http://wap.sciencenet.cn/blog-45849-1101009.html https://cunlab.org/2018/02/24/sclust_news/
write Nik-Zainal et al in the abstract of their 2012 Cell paper `The life history of 21 breast cancers’. The key figure of their paper shows a phylogenetic tree of tumor development in a patient. The paper contains lots of computational work on analyzing and interpreting mutations based on deep-sequencing data, but –a big surprised but— the very last step of putting together the tree was done manually. Half the paper is describing the reasoning that Peter Campbell and his group used to condense all the evidence they had gathered from genomic data into the tree – but there is no algorithm.
Obviously I was getting terribly excited when I saw this and started muttering to myself `I can automate this! I can put together an algorithm to build trees! That’s what I’m good at!’ Little did I know how difficult the problem was, how short the reads and how sparse the mutations. But my enthusiasm, if not my ignorance, must have been shared all over the world by all other computational biologists working in cancer genomics, because the last 1-2 years have seen quite a variety of approaches to assess tumor heterogeneity and reconstruct phylogenies.
Describing cancer as an evolutionary process is not a new idea, going back at least to Nowell 1976. Mutations are thought to increase the fitness of cancer cells and make the population grow faster, out-competing normal cells and other less-fit cancer cell populations. Cancer evolution has already been widely reviewed before the technological advances in sequencing over the last few years brought renewed vigor to the field.
Cancer evolution is currently a very active field of research and will be for a while. This is why I want to use a series of posts to discuss some of the basic underlying concepts and ideas.
A toy example of cancer evolution
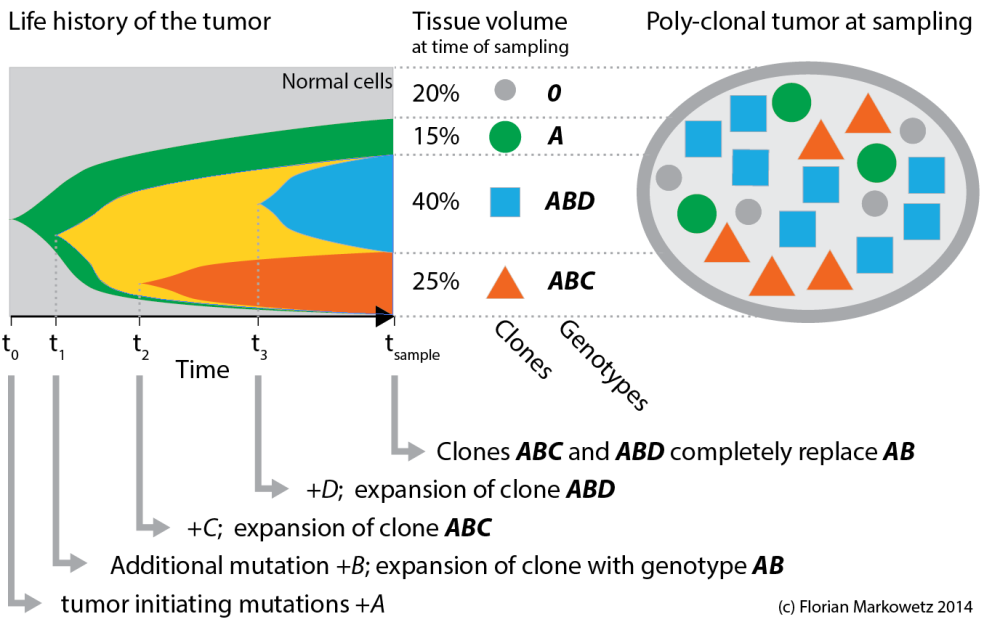 Figure 1. A tumour evolution toy example. A series of genetic changes (A, B, C, D) and expansion of clones characterized by them (A, AB, ABC, ABD) leads to a poly-clonal, genetically heterogeneous tumor by the time of sampling.
Figure 1. A tumour evolution toy example. A series of genetic changes (A, B, C, D) and expansion of clones characterized by them (A, AB, ABC, ABD) leads to a poly-clonal, genetically heterogeneous tumor by the time of sampling.
Let’s start with the toy example in Figure 1 above, which conceptually summarizes the evolution of a tumor from the first tumour initiating mutations to the heterogeneous tissue at the time of sampling (it is similar to Figure 7 in Nik-Zainal et al.).
The tumour sample on the right contains some normal cells (grey circles) and three different cancer clones (i.e. cancer cells that share a genome). Each clone is characterized by sets of mutations the cell population acquired over time (indicated by the letters A, B, C and D). Early mutations (A) are shared by all clones, while later mutations (C, D) differentiate younger clones. (I am using italic letters for mutations and bold italics for clones.)
The evolutionary process leading to this heterogeneity is summarized in the left plot, where colors correspond to clones. The shapes show the expansion of clones over time. At the time of sampling 3 clones are still in the tumour (*A*, *ABC*, *ABD*), while a fourth one (*AB*) has been replaced by its descendents.
This cartoon is obviously very simplified! For example most tumours will have many more very small clones. But even this simple toy example is already interesting enough to make some important observations.
A tree of clonal evolution
 Figure 2: A clonal evolution tree.
Figure 2: A clonal evolution tree.
The cellular composition of the tumour and the evolutionary relationships between the different components can be summarized in a tree, like the one you see in Figure 2 on the right. The numbers in the nodes correspond to the percentage of cells in the sample that belong to this particular clone. The grey top node indicates the 20% normal cells in the sample. The oldest clone is the green clone *A*, which is represented by 15% of cells in the sample. In the *A* population B mutations appeared and formed the *AB* clone, which is not present in the sample (0%) because its descendents *ABC* (25%) and *ABD* (40%) replaced it.
The way I have drawn the tree is also very simplistic and, for example, does not scale the edges according to mutation rates or evolutionary time passed. But as we will see (in later posts) even inferring such a simplified representation from data can be tricky!
This little example is already quite instructive. For example, we can see the difference between the frequencies of clones (*A* 15%, *ABC* 25%, *ABD* 40%) and the frequency of the mutations characterizing them (A 80%, B 65%, C 25%, D 40%), which are the sums of all cellular frequencies carrying this particular mutation. We can also see that ancestors and descendents can co-exist, which means that (some of) the inner nodes in the tree are populated. Finally, cancer trees have a well-define root node (the normal cells without mutations) and generally have a clear directionality, because mutations accumulate over time with child nodes keeping the parent mutations and adding more to them.
Infinite sites assumption. As you can see a key assumption underlying the analysis of mutation data is that each that each mutation appears only once and furthermore that once it appears, it does not revert back to its original state. This is called the infinite sites assumptions and from it follows that tumor evolution is a process of accumulation of mutations: earlier clones have less mutations and later clones have the early mutations plus some more.
The intra-tumour phylogeny problem
Using genomics technologies we can sample (features of) the genomes of the heterogeneous cell mixtures that is a tumour. These features can include single nucleotide variants, copy-number aberrations and CpG methylation.
The intra-tumour phylogeny problem is to infer a phylogenetic tree like the one in Figure 2 from this genomic sample. The problem comprises two sub-problems:
- Identify clones (= the nodes in the tree). If your data comes from deep-sequencing a mixed population of clones you will have to deconvolute this mixture to identify the clonal genomes. And if you have genomes of single cells from the tumour, you need to cluster them into clones.
- Relate the clones to each other (= the edges of the tree). Once you have the nodes you need to connect them in a graph, where they can take inner nodes and leaf nodes.
These tasks can be solved sequentially or jointly, and in the following posts I will discuss different methods to solve the intra-tumour phylogeny problem.
Quick recap: Last time we talked about tumor evolution and I presented a toy example to introduce key concepts. I also introduced the intra-tumor phylogeny problem: Given a sample of the genomes of clones in a tumour, reconstruct its `life history’. This problem consists of two sub-problems: (1) identification of clones, and (2) inferring evolutionary relationships between clones.
This problem falls into the general area of reconstructing phylogenetic trees — so how does inferring clonal trees compare to classical phylogenetic methods?
Classical phylogenetic trees
Joseph Felsenstein’s book Inferring Phylogenies is a classic in the field and the methods he describes have all been used in cancer genomics (eg here and here in one of my own papers). And Navin and Hicks show (conceptually) in a review from 2010 how different evolutionary scenarios can lead to different phylogenetic trees, so these methods could be very useful in differentiating between different theories of tumour evolution. Here is a link to their paper’s main figure with modes of evolution (a-e) and reconstructed phylogenies (f-j).
Clonal evolution trees
However, there are several reasons I believe that classical phylogenetic trees are not the best representation of clonal evolution in tumours. To start the discussion, the following figure compares classical phylogenetic trees to clonal evolution trees in the example we discussed in the last post.
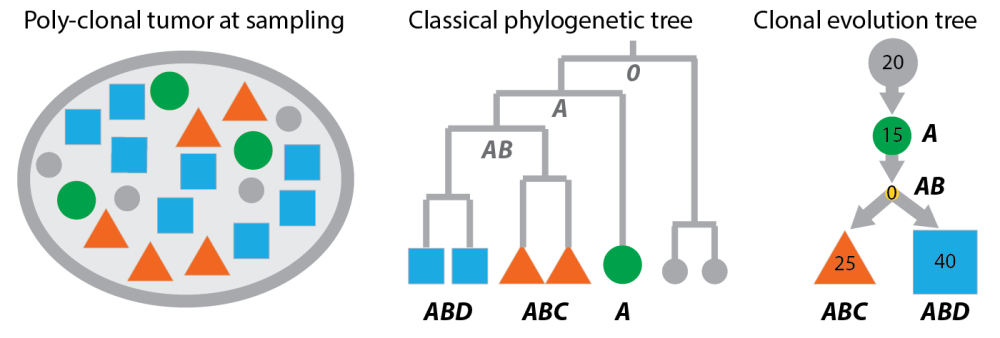 Classical phylogenetic trees compared to clonal evolution trees. Left: the poly-clonal tumour from the last post. Middle: cells sampled from the tumour arranged as leaves in a phylogenetic tree. The bold letters are the genotypes of the cells (according to the example in the last post). The grey letters in the tree are inferred ancestral genomes. Right: A clonal tree representation of the same tumour where nodes are clones (not cells) and inner nodes can be populated.
Classical phylogenetic trees compared to clonal evolution trees. Left: the poly-clonal tumour from the last post. Middle: cells sampled from the tumour arranged as leaves in a phylogenetic tree. The bold letters are the genotypes of the cells (according to the example in the last post). The grey letters in the tree are inferred ancestral genomes. Right: A clonal tree representation of the same tumour where nodes are clones (not cells) and inner nodes can be populated.
Now, if you compare the panels of this figure, you will find:
- Classical phylogenetic trees do not infer clones, but place the taxa –in our case observable feature of the tumour like single-cell genomes or methylation patterns– as leaf nodes in the tree. In classical phylogenetic analysis you don’t need to cluster your observations, because you have the Mouse genome and compare it to the Human genome, instead of multiple individual genomes of mice and men.
- Inner nodes of classical phylogenetic trees are unobserved. The ancestral genomes at these inner nodes can be inferred in a second step. However, in a tumour ancestors and descendants can co-exist so the tree representation should allow inner nodes to be populated (like the right panel of the figure).
- Classical phylogenetic trees encode distances between taxa, and because distances are symmetric the trees are undirected. However, the accumulation of aberrations in cancer genomes gives clonal trees a directionality: The child nodes carry the parent aberrations plus some more. Clonal evolution is more about asymmetric subset relations than symmetric similarities.
- The problems classical methods face in a cancer setting become even more evident when thinking about data from deep-sequencing a mixed population of clones. Without deconvolving this mixture, the taxa are not even defined and there is nothing you can put into the tree.
The need for new methods
There are some caveats to what I just said:
- You can cluster the leaves into clones by cutting the tree at different levels like in hierarchical clustering;
- While inner nodes are indeed never observed, edge lengths can be very small (or even zero) and thus effectively place leaves in the middle of the tree. For the plot above I (realistically!) assumed there was noise in the measurements of each cell. For perfect data, in contrast, the phylogenetic tree would have many edges of length 0, which means: the cells from each clone would have no distance between them and clone A would come to lie on top of an inner node — which would result in a phylogenetic tree pretty similar to the clonal tree I drew.
- Not all phylogenetic methods are distance-based and others like maximum parsimony or maximum likelihood might be more effective for cancer studies. And using an outgroup can help to establish directionality in any tree. But models of DNA evolution are generally time-reversible, whereas in cancer we can assume that somatic mutations don’t go away again, so I think there is still a difference here, no matter how you look at it. Working in directed models also has computational advantages and, for example, allows us to avoid time-consuming marginalization steps for the inner nodes.
So, in summary, even if the classical approaches are not a perfect fit for tumor evolution, they might come close enough in some cases. How well they do against methods directly built on principles of tumor evolution is the topic of on-going research (Edith and Ke will soon have something to say about that.)
Importantly, problem (4) stands tall and strong: cancer studies are mostly done on a mixed population of cells, which needs to be deconvoluted prior to evolutionary analysis.
 Figure 1: clonal evolution tree (details in previous post)
Figure 1: clonal evolution tree (details in previous post)
In the first post in the series I described a simple toy example to illustrate key concepts of tumour heterogeneity and evolution. A quick summary of the population composition and evolutionary relationsships is displayed in Figure 1 on the right. There are three clones present in the sample A, ABC, ABD characterized by four sets of somatic mutations A, B, C, D.
Our first discovery, when discussing this simple example in the last post, was that classical phylogenetic approaches might not capture important features of cancer evolution. So, which other methods are there to understand the evolution of clones in a tumour?
Principles of inferring tumour evolution
In this post and the next I want to discuss analysis approaches proposed in the last couple of years. Figure 2 organizes research strategies along basic principles, and this post (together with the next one) will discuss examples of each strategy in more detail.
- The first principal question is: does your method work on data from a mix of different clones (most of them do) or does it work on single cell genomes?
- The second question is: do you know which mutations appear together in the paternal or maternal copy of DNA (‘phased’), or do you have information of individual mutations without knowing the connection between them (`un-phased’)?
- Finally, does your method infer evolution from a single tumour sample or by integrating information from many? If you can do it from a single sample, then extending it to many is often straight-forward; but if you need many, you will often not be able to do anything with a single sample.
 Figure 2: Strategies to infer tumour evolution organized by basic questions.
Figure 2: Strategies to infer tumour evolution organized by basic questions.
Only five entries are filled, because I don’t know a method to infer the evolutionary history of a single cell. I will start by discussing 1a and 1b in this post.
1a: Single mixed sample, unphased
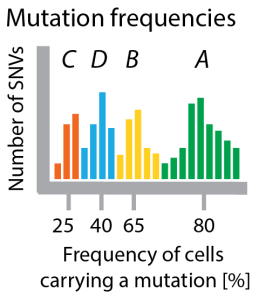 Figure 3 Clustering mutation frequencies hints at population structure of tumor. Bars show how many mutations appear with a certain frequency. I colored the clusters according to which clone in the tree they are specific for. In a real application you would not have this information.
Figure 3 Clustering mutation frequencies hints at population structure of tumor. Bars show how many mutations appear with a certain frequency. I colored the clusters according to which clone in the tree they are specific for. In a real application you would not have this information.
A prominent approach to sample the genomes of clones in a tumour is by sequencing deeply, such that many reads cover each mutation. In our running example this ideally should show that there are four (noisy) clusters of mutations, which are (on average) present in 25%, 40%, 65% and 80% of all cells (see Figure 2). In one of the first examples of this approach, Shah et al 2012 used deep sequencing to measure allelic abundance for 2,414 somatic mutations in triple-negative breast cancer.
Allelic frequency vs cellular frequency. One major challenge in these data is that in genomes as complex as cancer genomes the raw frequency of mutations (the allelic frequency) is not necessarily identical to the number of cells carrying the mutation (the cellular frequency, x-axis in Figure 3). This is why Sohrab Shah’s lab developed a method called PyClone, which uses mixture models to identify clusters of SNVs with the same frequency and at the same time corrects these frequencies for copy-number changes and loss of heterozygosity to estimate the fraction of cells in the tumor carrying these mutations. For copy-number data (instead of SNVs) similar methods exist, for example TITAN (also Shah lab) and THetA (from Ben Raphael’s lab).
Clusters are not clones. Each clonal genome is a combination of some of these mutation clusters. The number of clones is smaller or equal to the number of clusters. For example, there are only 3 clones in the tumour of our running example (A, ABC, *ABD*; AB died out), characterized by 4 clusters of mutations (A, B, C, D). The number of mutations in each cluster (the size of each hill in the histogram in Figure 3) have nothing to do with the number of cells that carry these mutations.
Clusters are not yet a tree. To relate the clusters to clones you need to order them in a tree. The clonal genomes are then given by the mutations that happened along the path in the tree to this node. For example, the orange triangle in Figure 1 represents clone *ABC*, because the path from the top (the grey normal cells) consecutively adds mutations A, B and C. To order the clusters into a tree, there are two approaches: either (1) first cluster and then build tree in an independent second step, or (2) joint clustering and tree building in an integrated model.
If you were to follow the first approach you could cluster mutations with PyClone and in our example establish four clusters with frequencies 25%, 40%, 65% and 80%. In a second step you could use this vector of frequencies as input to a method like TrApfrom Yuval Kluger’s lab. TrAp solves a highly constrained matrix inversion to reconstruct a tree consistent with the given frequencies. The tree in the ‘life history’ paper is also an example of the first approach, but shows limitations of consecutive clustering and tree-building: In their Figure 3D one of the clusters (called `cluster A’) had to be spread out over 3 positions in the tree, a discrepancy that could hopefully be avoided in an integrated approach.
Quaid Morris’ PhyloSub is an example of the second approach (actually the very first such example). They cluster the mutations with a similar mixture model as PyClone, but relate the parameters for each cluster in a tree structure (using what is called a tree-structured stick breaking process — which is too complex to cover here and hopefully will be the topic of another post.)
Clonal trees from SNV frequencies are generally not unique. Generally, reads are short and will almost exclusively only contain a single SNV, so the only information we have for tree building is the allele (or better: cellular) frequency. One of the first things Quaid and his team realized is that the trees you can reconstruct from frequencies are not necessarily unique. They identified several topological constraints. Take for example the toy tumour we have been discussing. Given the mutation frequencies of A (80%), B (65%), D (40%) and C (25%), a consistent tree could just order clones linearly by mutation frequency into:
A -> AB -> ABD -> ABDC.
This linear tree -incorrectly!- postulates the existence of four clones: A (15%), AB (25%), ABD (15%), ABDC (25%). (The way to compute this is: A exists in 80% of cells, B in 65%. If there is an AB clone, that leaves only 80-65=15% of cells for the A clone.) But given only frequencies of single mutations there is no way to distinguish this tree from the true tree in Figure 1.
SNV and CNA information can help each other: Many methods only look at one type of data, either SNVs or CNAs, but the combination of them can be very powerful. If you have a copy-number gain and you can find the same SNV on all copies you can infer that the SNV event was before the CNA. On the other hand if the SNV is only on one copy you can infer that the SNV was after the CNA (Kudos, Thomas).
 Figure 4 The first SNVs (in orange) can be found on all copies of a later amplification, whereas SNVs after the amplification (blue) are only found on individual copies. These observations help to establish an order between SNV and CNA events.
Figure 4 The first SNVs (in orange) can be found on all copies of a later amplification, whereas SNVs after the amplification (blue) are only found on individual copies. These observations help to establish an order between SNV and CNA events.
1b: Single mixed sample, phased
Depending on technology reads can be longer and span more than one mutation. The long-read example I know best is Sottoriva et al (2013), who present methylation data from 454 sequencing of the IRX2 molecular clock, a 201bp locus on chromosome 5, which spans 8 CpG regions (potential methylation sites). Every read can be represented as a binary pattern of length 8 (where 1 is methylated and 0 is unmethylated). That is much more information than in the examples above, where every read only carries the information ‘there is an SNV’ or ‘there is no SNV’.
Methylation data has the added advantage that it’s error rate is 10,000-fold higher than that observed for nucleotide substitutions, which gives it a much higher resolution as a marker of cell fate.
The major drawback, if you can call it that, is that methylation is a reversible process, whereas for SNVs you can safely assume that they don’t back-mutate. Thus, for SNVs you see an accumulation of events during tumor development, which makes it easier to infer a direction of the process, which is much harder for methylation (and needs further, often artifical, assumptions, like normal tissue being completely unmethylated).
Our own -still unpublished- approach to infer tumor evolution from methylation data is called BitPhylogeny (for Bayesian intra-tumor phylogeny) and just like Quaid Morris’ PhyloSub it uses a nested stick-break process to sample trees for a mixture model. The code is available at https://bitbucket.org/ke_yuan/bitphylogeny — feel free to try it out, we are happy about any feedback. I will post more details in a future post.
2a: Multiple mixed samples, unphased
This section conceptually focuses on how to infer cancer phylogenies from single nucleotide variants (SNVs) identified by deep-sequencing a cancer genome.
Modeling multiple samples. From a statistical perspective you need to specify a probability for the state of each SNV in the sample given its cellular frequency: Pr( sample | cf ). For multiple samples, the simplest assumption is assume data (reads counts) from different samples to be conditionally independent given their cellular frequencies:
Pr ( sample1, sample2, cf1, cf2 ) = Pr( sample1 | cf1 ) Pr( sample2 | cf2 ) Pr( cf1, cf2 ).
Now how to specify Pr(cf1,cf2)? Methods like PyClone assume independence. But in the case of serial blood samples or circulating tumor DNA from plasma there will be temporal dependencies, which you might want to model. And different tumor sites might exhibit spatial dependencies. So if you are really into modelling you might want to put everything together in a big spatio-temporal model. Kriged Kalman-filters anybody?
I plan a future post specifically about details of statistical modeling; the remainder of this one will be more about basic ideas and concepts.
 Figure 2: The clusters with frequency 25%, 65% and 80% are found in both samples, but sample 2 shows that the 40% cluster in sample 1 actually consists of two separate clusters, which just happened to sit on top of each other.
Figure 2: The clusters with frequency 25%, 65% and 80% are found in both samples, but sample 2 shows that the 40% cluster in sample 1 actually consists of two separate clusters, which just happened to sit on top of each other.
Multiple samples can help identifying SNV clusters.Last time we discussed that one of the first steps in analyzing deep sequencing data from a tumour is to cluster SNVs by frequency, ideally correcting for copy-number changes and LOH, to get clusters which (on average) appear in the same number of cells (i.e. have the same cellular frequency).
Now what if you are unlucky and there two groups of SNVs which both happen to appear with a frequency of 40% each, but in different cells. In the frequency distribution these two sets would sit right on top of each other and you would not be able to distinguish between them.
This is where a second sample can help. If you were unlucky with the first sample, you might be lucky in the second one and the two clusters might appear with different frequencies. Figure 2 shows an example.
Multiple samples can show the progression of disease. Here I am considering a toy example where a cell from a minor subclone in one tumor (think of the primary tumor or a pre-treatment tumor) seeds a second tumor (a metastasis or a post-treatment tumor). Figure 3 has hypothetical clonal evolution trees. You see that the one cell carries its evolutionary history with it. The second tumor doesn’t start `from scratch’ (from the normal tissue) but from an already mutated cancer genome, to which it adds even more mutations.
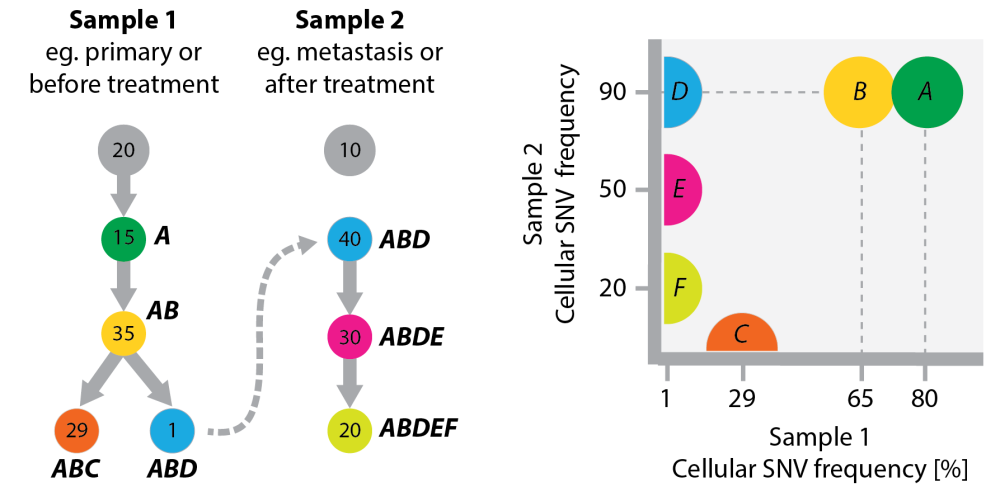 Figure 3 Left: A toy example of a clonal tree in two samples from the same patient, which are linked because a cell from a subclone in sample 1 gave rise to sample 2. Numbers in the tree are frequencies of clones. Tumor evolution in sample 2 does not start from normal tissue (thus no arrow out of the grey node at the top). I have included a grey circle in both samples to indicate that both will have normal contamination. Both grey circles can represent the same tisse, eg if the samples are before/after treatment. Right: The plot on the right compares the cellular frequencies of SNVs between the two samples. The half circles at the axes indicate SNV clusters that (i) either don’t exist in one of the samples (C,E,F) or (ii) are below the detection limit (D).
Figure 3 Left: A toy example of a clonal tree in two samples from the same patient, which are linked because a cell from a subclone in sample 1 gave rise to sample 2. Numbers in the tree are frequencies of clones. Tumor evolution in sample 2 does not start from normal tissue (thus no arrow out of the grey node at the top). I have included a grey circle in both samples to indicate that both will have normal contamination. Both grey circles can represent the same tisse, eg if the samples are before/after treatment. Right: The plot on the right compares the cellular frequencies of SNVs between the two samples. The half circles at the axes indicate SNV clusters that (i) either don’t exist in one of the samples (C,E,F) or (ii) are below the detection limit (D).
The right-most panel in Figure 3 compares the SNV frequencies between samples from the two tumors (given the cellular frequencies of clones annotated in the trees).
- Early SNVs (A and B) sit in the top right because they appear in almost all cells of both samples.
- SNV D only appears in 1% of cells in tumor 1 and is thus below the detection limit of most current sequencing technologies, whereas in tumor 2 it appears in all cells. If you don’t know the evolutionary history of the samples you could explain these SNV frequencies in two ways: (i) predict a minor sub-clone in tumor 1 or (ii) assume that an *AB* cell seeded tumor 2 and D was a very early event in tumor 2 development. In both cases you had evidence to conclude (correctly) that D (but not necessarily A and B) is important for the transition that happened between tumor 1 and 2. You need data from both tumors for this conclusion. Had you only seen data from tumor 2, then A, B, and D would be undistinguishable because they appear all with the same frequency.
- If you don’t know the temporal order between the two samples, you could use the SNV data to infer it. The fact that D, E and F were found in tumor 2 but not tumor 1 would indicate that tumor 2 developed out of tumor 1 (cancer accumulates mutations). However, alternative branches in tumor 1 development (like C) can complicate this approach.
Multiple samples can help to find forks in trees. In Jiao et al 2014, Quaid Morris and his team describe topological constraints for evolutionary trees. I have summarized their examples in this table (using the numbers from their Figure 1):
 The table illustrates three principles of building phylogenetic trees from SNV frequencies:
The table illustrates three principles of building phylogenetic trees from SNV frequencies:
- With only a single sample, clones can always be ordered linearly (Sample 1).
- They can also be arranged as a fork, unless the sum of frequencies of child nodes is larger than the frequency of the parent node (Sample 1′; B+C = 60%+40% > 80% = A). Quaid calls this the ‘sum rule’. The same idea is called the pigeon hole principle in Nik-Zainal et al (2012) and is also implied in the constraints used by Strino et al (2013). While this constraint applies already to a single sample, additional samples increase the chance to be able to use it.
- The last constraint is specific to multiple samples: Quaid and his team describe the ‘crossing rule’ where a change in frequencies between two samples can only be explained by the clones sitting in independent branches (Sample 1+2). In this example here there would be two linear chains: A -> B -> C in sample 1 and A -> C -> B in sample 2, which is a contradiction to the assumption that we observe the same evolutionary process in both samples. The only possible solution is to conclude that B and C live in separate branches of a fork below A.
The pigeon-hole principle also entails that if two separate clusters indeed sit on top of each other (like in Figure 2) their cellular frequencies must be below 50%. Else there would be a cell carrying both SNVs and the two clusters were in fact only a single cluster.
Multiple samples can be the nodes of a tree. Another advantage of having several samples per tumour/patient is that you can use them directly for tree-building, even if the resolution of the data is not high enough to reveal the clonal composition of each sample. In Schwarz et al (2014) we develop a distance-based approach to infer a phylogenetic tree from copy-number profiles of multiple tumour samples.
Comparing copy-number profiles is challenging (and in particular much more challenging than counting SNVs) because
- copy-number aberrations come in all sizes. Comparing two genomes base-by-base without taking the size of aberrations into account (in technical jargon: the horizontal dependencies) can give you a completely wrong estimate of how many changes happened between two genomes.
- copy-number aberrations can show complex cascading and overlapping patterns, which makes counting the number of changes even harder.
In our paper we show how to use finite-state transducers to tackle these two problems. This is a machine that runs along two genomes to `translate’ one into the other and which in the process counts the minimal number of changes required to do so. The method is called MEDICC for Minimum Event Distance for Intra-tumour Copy-number Comparisons.
Our approach also phases copy-number variants by assigning them to one of the two physical alleles such that the overall evolutionary distance is minimal (this is a heuristic, but it works). We also introduce summary statistics of tumor evolution that can be used (and are being used in a so far unpublished follow-up paper) to link tumor evolution to patient outcome.
If you are interested in these questions and methods, check out the software here and the paper here.
The following figure shows an example of a tumor evolution tree for a patient with endometrioid cancer.
Figure from Schwarz et al (2014): Phylogenetic Quantification of Intra-tumour Heterogeneity. The tree shows evolutionary relationsships between 18 samples of an endometrioid cancer.
2b: Multiple mixed samples, phased
Section 2a turned out to be longer than expected, so it might just as well be a good thing that the only paper I can right now think of to discuss here is Sottoriva et al (2013), which was already in the last post.
I am happy about all suggestions what other papers and approaches fit into this section.
That’s all for now.
Welcome back to the intra-tumour phylogeny problem. Let’s take a quick breather and see what we have got to so far:
- Introducing the intra-tumour phylogeny problem;
- Comparison to classical phylogeny;
- Methods for single samples;
- Methods for multiple samples.
And today’s topic finally is:
Single cell analysis
Single cell sequencing wherever you look!
In breast cancer (e.g. here and here). In leukemia (e.g. here and here). And some very visible studies from the BGI in renal carcinoma, a myeloproliferative neoplasm, bladder cancer and colon cancer. That’s certainly enough material to start reviewing it.
“Cancer genomics: one cell at a time” by Nicholas Navin gives a very good overview of methods to isolate single cancer cells, amplify their genomes, profile mutations and reconstruct evolutionary trajectories. And -even better- the review goes beyond a simple laundry list of methods to comment on their strengths and limitations. If you are interested in single cell genomics in cancer, this is a must-read.
I had originally planned to write a more methods-focused post (on what you actually do with all those genomes), but this will have to wait and here I will use Navin’s review as a starting point for my own discussion of some conceptual points that went through my mind while I read it:
- Cells are part of a tissue (Not really a big surprise, I hope, but something often forgotten in genomics studies);
- Phylogenetic methods will have to be adapted for single cell data (yes, you have seen that idea before);
- Single cell genomics will not completely replace bulk-sequencing for clinical applications (which means that all the stuff we have discussed so far will stay important).
Cells in context: In situ analysis
“Biologists have been studying single cancer cells since the invention of the microscope by Antonie van Leeuwenhoek in 1665.
Many initial observations were based on the morphological differences between tumor cells, as recorded in the late 1800s by early pathologists, such as Rudolf Virchow.” (Navin, 2014)
This is an important point. In a technology-driven field like genomics, it’s particular important to highlight the long history most research questions have.
“These observations were greatly improved by the development of cellular staining techniques, such as hematoxylin and eosin (H&E).
In the 1980s, the development of cytogenetic techniques, including spectral karyotyping (SKY) and fluorescence in situ hybridization (FISH), galvanized the field by allowing researchers to visualize the genomic diversity of chromosome aberrations directly in single tumor cells.” (Navin, 2014, references removed, Wikipedia links added)
Staining techniques like FISH give you a glimpse of single cell genomes. And even more: you can look at the spatial context of each cell because the staining is in situ. Are the same aberrations shared by neighboring cells or do they differ? These are important questions about the location and spread of cancer clones that can only be answered by looking at cells in their tissue context.
And, what’s even better, you can deduce tumour phylogenies from FISH data; see for example Russel Schwartz’s work on modeling copy number changes (PMID 25078894, 23812984)
But Navin continues:
“However, only in the past four years has the field moved from qualitative imaging data to quantitative datasets that are amenable to statistical and computational analysis.” (Navin, 2014)
No, I’m afraid, I don’t completely agree with this.
This sentence is from the beginning of the review and not at its heart — but calling genomics quantitative and imaging qualitative bothers me enough to comment on it.
Images can be as quantitative as genomes – it really all depends on how you analyse them. You can use computation and statistics on image data as easily as on genomes. Here is an example from my own lab: by analyzing standard pathology H&E slides we showed how quantitative image analysis of cellular heterogeneity in breast tumours complements genomic profiling.
And H&E is not the only example. Kornelia Polyak’s lab used a combination of immuno-fluorescence with FISH (called IFISH) for the quantitative analysis of genetic and phenotypic features and their spatial distribution in breast tumours.
To make the analysis even more quantitative, Anne Trinh in my group has developed GoIFISH for the quantification of genomic alterations and protein expression obtained from IFISH data (the paper appeared in the same collection as Navin’s review). We are currently using GoIFISH on larger patient cohorts – more on this once we have results.
So, in summary, the situation is this: either you get spatial information about the tissue (but only for a few markers and not the complete genome) or you get the genomes of individual cells but lose the spatial information of where the cells sit in the tissue. Tissue and genome are important, but with current technologies you can’t have both at the same time.
Single cell genomics
Ok, let’s move on to the next part of Navin’s review, which highlights the importance of single cancer cells in tumour initiation and progression:
“While there is substantial evidence that tumor cells can communicate with their neighbors and the stroma, there are also many complex biological processes that occur through the actions of individual cancer cells.
These processes include the initial transformation event in a normal cell, clonal expansion within the primary tumor, metastatic dissemination and the evolution of chemoresistance” (Navin, 2014; emphasis added)
Now this is the second thing I am not completely happy with. Navin refers to the “substantial evidence” only to dismiss it again immediately.
And there is some ambiguity: what does “occur through the actions of individual cancer cells” really mean? Does it mean (1) “involving individual cancer cells (plus other factors)” or does it mean (2) “involving only an individual cancer cell without any other important factors”.
I think (1) is the correct answer:
- Initial transformation: True, it starts with one renegade cell, but the new tumour has to compete with the normal microenvironment to overcome antitumourigenic pressures, which is one of the reasons why we don’t get more cancer. Thus, already the first steps of tumour formation are multi-cellular processes and do not occur just through the action of an individual cancer cell.
- *Metastatic dissemination: *Individual cancer cells that are shed from the primary tumour in search of a new niche to colonize are obviously an important part of metastatic spread, but they are certainly not the only important factor! For example, not every cancer can metastasize to every organ, because it needs favourable tumour-stroma interactions (the so called seed and soil hypothesis). Thus, metastatic dissemination is a multi-cell process involving both cancer and normal cells.
- Clonal expansion within the primary tumour: I hand it over to the Polyak lab again. In a recent paper they show that non-cell-autonomous mechanisms can drive tumour growth. A less fit, small clone can support the growth of the whole tumour and when it gets outcompeted by faster proliferating competitors the tumour collapses. Thus, clonal expansion is a multi-cellular process.
None of these points argues against looking at the genomes of single cells – but, please, let’s not forget about the context in which these genomes matter.
Single cells in the clinic
“In the near future, [single-cell sequencing] will begin to be applied to the clinic in early detection, prognostics, diagnostics and therapeutic targeting and thereby will have a direct impact on reducing morbidity in many human cancer patients.” (Navin, 2014)
I hope this is true. Everything that helps patients is good.
But what does “in the near future” mean? All histories of cancer research I have ever seen only allow one conclusion: If they have started to work on it now, there might be individual clinical research projects with results in 5 years, and whole programs in 10, but routine use outside of cutting-edge cancer centers will be more than 20 years from now. And that all depends on single cell genomic techniques to prove themselves superior to existing methods.
From a basic biology perspective I am all for single cell studies. In theory they should allow the most highly resolved view of genetic heterogeneity in a tumour. And for me this is already enough motivation to look at these data – not everything has to be translational.
But for clinical use, I am not sure we have even started to exploit the information we can get from sequencing mixed tissue biopsies. As you know from the last posts in this series, there is a lot of information in mixed samples.
And there will always be tissue biopsies until we have understood much better how the individual tumour cells and DNA fragments that circulate in the blood (and that are marketed as liquid, noninvasive biopsies) get there, what biases they have, and how much they can actually tell us about the tumour.
There is still a lot of hard work to be done until we can test whether single-cell methods are more accurate in finding actionable mutations in minor sub-clones and lead to better treatment at the same cost as sequencing bulk tissue.
A cancer is more than the sum of its cells
Just to be clear: I am not arguing that single cell sequencing is not informative or important. It certainly is!
I just don’t want their importance being oversold. Single cell techniques are the newest and hippest kid on the block, but that doesn’t mean everything else is outdated. And we still have a long way to go until single-cell studies reach peak performance (which Navin (2014) describes very well):
- Technological improvements need to reduce measurement biases and improve overall data quality.
- Better algorithms need to bring down the error rates in calling mutations in single genomes.
- Additionally: All the single-cell papers listed in the beginning use classical phylogenetic methods for inference of clonal evolution, even though (i) they work on cells, not clones, (ii) they are undirectional, and (iii) inner nodes are by definition unobservable – all of which don’t hold in clonal evolution.
Tumors are more than collections of individual cells. But cellular interactions and the influence of tissue architecture are generally ignored in genomics studies.
This is really a limiting factor for evolutionary analysis. After all you cannot talk about the fitness of clones and ignore the environment for which they have to be fit for.
What we really want in the future is a genetic and phenotypic 3D model of a tumour: Which cell with what mutations sits next to which other type of cell? For example: Does mutation X only appear in lymphocyte-rich parts of the tumour? Also: What morphologies do cells with that particular mutation have? Or other similar questions.
Single-cell sequencing is great, but only the first step to a comprehensive characterization of tumours which needs to combine genomes with spatial tissue organization. No single technology will allow this comprehensive view and the future goal will be to integrate the limited snapshot each individual technology can provide.
Long way to go …
Florian
Acknowledgements: Thanks to Edith Ross for a rigorous review of early drafts of this post.
Some references
Navin, N. (2014). Cancer genomics: one cell at a time Genome Biology, 15 (8) DOI: 10.1186/s13059-014-0452-9
Yuan, Y. (2012). Quantitative Image Analysis of Cellular Heterogeneity in Breast Tumors Complements Genomic Profiling Science Translational Medicine, 4 (157), 157-157 DOI: 10.1126/scitranslmed.3004330
Trinh, A. (2014). GoIFISH: a system for the quantification of single cell heterogeneity from IFISH images Genome Biology, 15 (8) DOI: 10.1186/s13059-014-0442-y
What do you picture when you hear the word ‘clone’? A white-clad imperial stormtrooper from Star Wars: Attack of the clones? Or a fluffy sheep called Dolly? Both are good choices. Both are good, solid, well understood clones. But how is the situation in cancer? This is where it gets difficult. In most talks (at least the ones I sit in) the word ‘clone’ is used very loosely like it was a trivial concept. My goal for today is to show that reality is more complex than the ‘plain vanilla’ version that is often described on some introductory slide.
I found some interesting comments to one of my recent posts trying to explain why ‘real’ evolutionary biologists have traditionally not been that interested in cancer. Erick Matsen wrote:
I was a little put off looking into the area when there was a recent pile of papers working on various ways to define clones (…). I’m not sure if I’m interested in entering such a “hot” field.
And David Posada wrote:
[L]ack of individuals: evolutionary inference is often made on populations of individuals, or on individuals from different species. Until the appearance of single-cell genomics (..) cancer data was on pooled individuals, which makes, in my opinion, evolutionary inference more complex and less powerful.
“Various ways to define clones” … “lack of individuals” … they have both spotted a key problem of cancer evolution studies: most data are from bulk sequencing a single tumor sample which pools all the different cells and clones in there. The ‘populations of individuals’ are the clones, but they are not a priori defined and need to be reconstructed from the mixture (see the initial posts in this series).
Inferring clonal evolution is indeed a hot field. As hot as it gets, actually. The ICGC pan-cancer project has a whole working group dedicated to the inference and characterization of tumor clones in 2500 bulk-sequenced samples. And a DREAM challenge on tumor phylogenies from bulk samples will soon open shop.
So, with all that activity on clonal evolution, do we at least understand well what a clone is?
The simple case: clonal and sub-clonal aberrations
Let’s start the discussion with individual aberrations (mutations, copy-number changes, basically anything you can do to a genome). An aberration is called clonal if it appears in all cells of a tumor. If it appears in fewer cells, it is called subclonal.
Here, clonality is a statement about cellular frequencies: 100% = clonal; <100% = sub-clonal (assuming you have already corrected for the number of normal cells in the sample). A popular way to assess clonality of a mutation is clustering by SNV frequency as we discussed in earlier posts.
Clonality is also a statement about the order of appearance: if an alteration is clonal, we take this as evidence that it was already there in the very beginning of the tumour, whereas sub-clonal aberrations are thought to appear later in tumor development.
What do we talk about when we talk about a clone?
So much for aberrations. Now let’s talk about cells. A popular definition is the following:
[A clone is a] set of cells that share a common genotype owing to descent from a common ancestor.
In some contexts a clone is more restrictively defined as a set of genetically identical cells. (Merlo et al 2006)
Problem 1: clusters of mutations are not yet clones
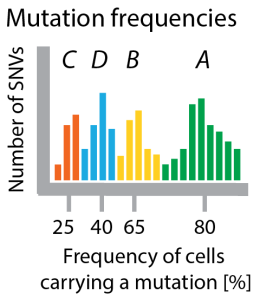 Figure 1: A cartoon histogram of SNV frequencies showing four clusters. These four clusters are not necessarily four clones. Depending on their frequency and phylogeny there might be less clones than clusters.
Figure 1: A cartoon histogram of SNV frequencies showing four clusters. These four clusters are not necessarily four clones. Depending on their frequency and phylogeny there might be less clones than clusters.
As a first step, let’s just accept this as a good definition. Then we are left with a practical problem: Linking the sets of mutations occurring at different (mostly subclonal) frequencies to sets of cells is not straight-forward. I have gone through this inference problem in more detail in a previous post.
Quick recap: to infer clones from clusters you need a phylogeny that tells you how the clusters relate to each other. For example, if you have one cluster at frequency 50% and one at frequency 30%, then only the phylogeny can tell you if there is a cell having both sets of mutations or if they live on separate branches. The frequency of clusters plus a phylogeny can tell you which cell populations with shared genotype (= the clones) exist in a tumor.
In practice, it is really hard to infer unique phylogenies if all you got are allele frequencies (this is what David Posada means when he says ‘complex and less powerful’) and a lot of uncertainty remains (see e.g. the PhyloSub paper for a discussion).
This means: while it is not too difficult to cluster mutations into sets of equal (allele or cellular) frequencies, making the additional step to predict the genotypes and frequencies of cell populations (= clones) is very hard. At least for bulk sequenced data, which makes up almost all the data that is out there.
Problem 2: there are no two cells with identical genome in the tumour
But it is even worse than that. Not only are there practical problems, I think there are conceptional problems too. The definition above talks about ‘cells sharing a genotype’ or ‘genetically identical cells’ –but do such cells actually exist? I don’t think so. Instead, I claim that with high likelihood no two cells in a tumour have a completely identical genome.
My thinking goes like this: The mutation rate of the human genome is notoriously hard to estimate, but I found some numbers for healthy tissue that give at least a first orientation: Bionumbers says 10^-11, Lynch 2009 says 10^-9 and Bozic and Nowak 2013 say 10^-9 to 10^-10.

Figure 2: Probability that genome stays identical
These little differences matter! You can see that by a back-of-the-envelope calculation how likely it is that the genome stays identical through cell division. Assuming that bases are independent and all have the same mutation rate, this probability is (1-mutation rate)^(number of bases). The figure on the right plots this value for different mutation rates for a genome of 3 billion bases. For a mutation rate of 10^-10 the probability of staying unmutated is 74%, for 10^-9 it is 5% and for 10^-8 it is pretty much 0.
In cancer we will be on the right side of this plot. First of all, we expect mutation rates to be higher than in healthy tissue, and additionally, copy-number changes and structural variation also contribute to the mutational load of a cancer genome, but are not covered in the mutation rates cited above.
As soon as the mutation rate is higher than a still healthy 10^-9 you can be pretty certain to see at least one mutation per cell division. This means: every cell in a tumor will have its own genome and there are no sets of cells with identical genomes. The more accurate sequencing technologies become, the more we will see of this diversity.
Defining a clone as a set of genetically identical cells sounds straight-forward until you realize that you will end up with as many clones as cells.
Problem 3: All cells in a tumor descent from one renegade cell
This leaves us with the last remaining part of the definition of a clone: The descent from a common ancestor.
Cancers are generally thought to start with one renegade cell. All the heterogeneity and genetic diversity we observe develop out of this cell, which is the common ancestor of all cells in the tumor. So if descent from a common ancestor is the criterion for being a clone, then the whole tumor is a single clone.
This might be the reason that some people speak of subclones instead of clones. The subclone is a part of the tumor clone. But which part? If you define it by genetic identity, you will run into the same problems as discussed in the last section.
What now?
Here are some ideas, which are modifications of the definition we discussed above. Figure 1 shows a small toy example of the history of tumor cell population. Circles are cells, colors correspond to clones, boxes are mutations.
 Figure 3: A tree following the development of a tumor cell population (A – G) from a normal cell. The cell population consists of three clones (red, green, blue), sets of cells with no mutation occurring between them and their most recent common ancestors (cells 1-3).
Figure 3: A tree following the development of a tumor cell population (A – G) from a normal cell. The cell population consists of three clones (red, green, blue), sets of cells with no mutation occurring between them and their most recent common ancestors (cells 1-3).
To define a clone you need to bring different ideas together:
- Genetically identical cells. No two cells might be genetically identical in a tumor with a high mutation rate, but not all these changes might matter. If we want to describe the structure of the population we could restrict mutations to a predefined set (e.g. only known drivers mutations) and then define a clone as a set of genetically identical cells based on only these markers. This will ensure that a clone is more than a single cell, but opens the door to arbitrariness how to select the marker set, something that apologists of genome-wide ‘unbiased’ approached do not like.
- Identical by descent: The reason for genetic identity of a clone should be that all cells are descendants of the same ancestor (the cells numbered 1-3 for the three clones in Figure 3). If a mutation can only arise once during tumor development then all genetically identical cells will live on the same branch of the tree, but if mutations appear more than once, there could be (but maybe not very likely) two clones with the same genomes arising two different branches of the tree.
- Maximality: By the definitions in items 1 and 2, {E,F,G} is a clone, but its subset {F,G} would also be a clone. To remove this redundancy we should demand that the set cannot be extended by other cells and still be a clone. Then only {E,F,G} is a clone and {F,G} is not.
This definition applies to cells that are in the current cell population at the time of sampling. I am not sure what to do about historical clones like the yellow one that once lived in the tumor, but not anymore. We see the mutations that defined them in all their descendants, but how do we define them? And do we need to?
I hope I didn’t make things more complicated than they already are. At least for me it was helpful writing all this down.
What makes a cancer deadly is not necessarily the growth at the location where it started (the primary tumour) but its spread through the body to other organs and tissues (called metastasis). Better understanding the metastatic process is one of main reasons we are interested in inferring cancer evolution.
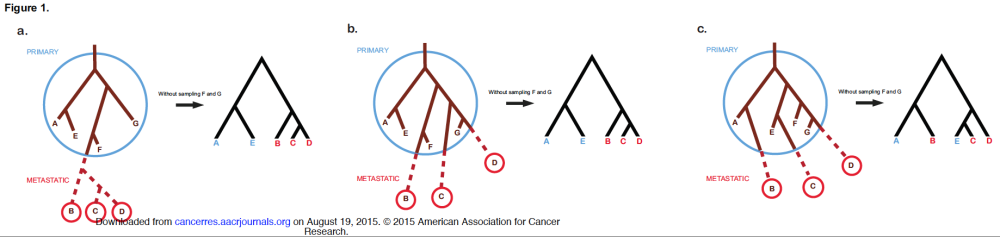
Today I would like to summarize and discuss two recent papers on cancer phylogenetics and metastasis. The first paper is the comprehensive review by Naxerova and Jain in Nature Reviews Clinical Oncology titled “Using tumour phylogenetics to identify the roots of metastasis in humans.” The second paper is an Opinion paper by Hong, Shpak and Townsend in Cancer Research titled “Inferring the origin of metastases from cancer phylogenies.”
Using tumour phylogenetics to identify the roots of metastasis in humans
Naxerova and Jain’s review has two parts. In the first part they describe the features of different models of metastasis, the second part describes different measurement techniques (histopathology, somatic copy-number alterations, single nucleotide variants, X-chromosome inactivation, CpG methylation, microsatellite analysis, whole genome studies — nice summary table here). I will focus on the models.
Models of metastatic spread
The systemic disease model assumes that there is no connection at all between different tumours in the same patient, which all arose independently of each other.
The linear progression model assumes that metastasis happens by a genetically advanced cancer cell late in the life of the primary tumour, which leads to a small genetic distance between primary and secondary neoplasm. If this happens multiple times you will get a star topology of metastatic spread (with short edges).
The metastatic cascade model additionally assumes that one metastasis can shed cells to start the next metastasis, which can lead to a more complex tree model.
The parallel progression model, on the other hand, posits that metastasis occurs early and that primary and secondary tumours develop independently, leading to high genetic distance and again a star topology (but this time with long edges).
And just to make things even more complex the self seeding model allows for cancer cells to come back to the primary, that is “bidirectional, dynamic cell exchange between synchronous lesions”. Uh oh, that means the graphs might get messy. In a standard phylogeny the clones you find in a sample form one subtree of the lineage tree, but if you allow self seeding then the samples can spread more than one subtree.
These different models are not mutually exclusive and a priori there is no reason why not early on a secondary tumour could start to develop independently while the primary is at the same time starting a metastatic cascade.
I have collected some features in this table:
| Model | genetic difference**primary-metastasis** | time of metastasis | phylogeny |
|---|---|---|---|
| Systemic disease | high (all independent) | NA | no edges |
| Linear progression | low | late | linear/star |
| Metastatic cascade | low (in mets); high (to primary) | late | tree |
| Parallel progression | high | early | star |
| Self seeding | lowering it | ? | cyclic graph of samples |
And as always with the Nature Reviews journals they have very well done overview figures like the one on the right to illustrate the different models. Click on the figure to go to the Nature Reviews Clin Onc page.
Challenges to inferring roots of metastasis
Importantly the review also identifies some challenges and limitations to inferring the roots of metastasis from patient samples. One of them is dormancy which can distort estimates of the age of tumours:
“[W]hether a metastatic lesion arose after a prolonged latency period because it disseminated late in cancer progression or because it underwent a period of dormancy at the distant site might be difficult to judge.” *
Another challenge is underestimating the clonal diversity of the primary tumour:
Because we cannot sample every single part of a primary tumour —in the clinical setting, analysing all of the tumour is de facto impossible because some parts are required for diagnostic purposes— one cannot exclude the possibility that the area harbouring the pre-metastatic clone was missed. *
Evolutionary theory to the rescue?
This directly brings me to the second paper by Hong, Shpak and Townsend (HST for short), who were trained as evolutionary biologists and comment on research in cancer evolution from their perspective.
They reiterate the issue of underestimating the clonal diversity of the primary tumour:
We argue that the chronology of metastatic events cannot be established without complete information on the phylogeny of subclonal lineages within the primary tumor. *
Their figure 1 shows how very different evolutionary histories can result in the same inferred phylogeny if not all clones in the tumour are sampled.
It is essential to have people trained in evolutionary biology start contributing to cancer research — because, as you know, the biggest problem in cancer evolution is that mostly people like me are doing it. (As an aside, I found it remarkable that the Nature gathering on cancer heterogeneity did not include a single evolutionary biologist — it shows the intellectual biases in the community.)
Now, what do HST have to offer? Experimentally their advise is to use one of two data gathering strategies:
- Sample a large number of small, spatially separated sections of the tumor (assuming that genetic diversity is spatially partitioned and each small portion is homogeneous)
- Perform single-cell sequencing of enough cells (HST acknowledge that it is not clear how many that might be).
HST explicitly criticize our ovarian cancer paper, “because of the absence of necessary information on spatially distinct subclonal heterogeneity within the primary tumor,” but failed to spot that we were actually following their approach number 1.
The problem is that the biology of ovarian cancer does not easily fit into the `localized primary and mets’ scenario HST discuss. At presentation most ovarian cancers are already spread throughout the lower body and there is little evidence where the tumour started. Clinically this spread-out tumour mass is often considered the primary tumour. So the different samples we used in our paper are not metastases (in the way HST think about it) but bits and pieces of the same “primary” tumour.
I understand this is not obvious to people not working in ovarian cancer and we could maybe have been more careful qualifying the word ‘metastasis’ in our paper.
What do HST have to offer on the methods side? Not very much. They don’t like the methods we use in the cancer field and would like to see them replaced by “well-established character-based phylogeny reconstruction methods based on molecular evolutionary models” without giving any detail on how that might look like.
In particular they don’t like our clonal expansion index:
It is not the null expectation for any population evolving along a phylogenetic tree, nor is it the null expectation for the distributions of genotypes of organisms in a population, all for the same reasons: genealogical relatedness, finite time, and spatial structure. *
I agree with them.
We thought of our work as the beginning of the story, not the end of it.
Please, show me how it’s done better!
My prediction is that HST will struggle to directly apply the “well-established evolutionary methods” they have been trained in to the complexity and noise of cancer genomics data. The evolutionary methods they will eventually discover to work well on these data will look very different from the textbook examples they had in mind while writing their paper.
HST have talked the talk, now let’s see how successfully they walk the walk.
癌症基因组里的clonal和subclonal的概念
癌症里的clonal演化概念由Nowell 1976年在Science里提出来的(Science. 1976 Oct 1;194(4260):23-8.),他假设人体的某个体细胞发生突变后经历了数个clonal扩增(演化)过程,最终这个含突变的体细胞集群就演化成了癌组织。这个过程和群体遗传学的溯祖理论(colescent)的祖先想,以后的colnal 演化模型借鉴共同祖先这个概念不是溯祖理论本身。clonal被广泛翻译“克隆”。我觉得这种译法不妥当,因为这里的clonal指的的带突变的细胞群落。所以,我这里把clonal译成集群,subclonal译成子集群比较合适。
随着二代测序技术的发展,这个假说经历了漫长的沉寂期后在2008s左右迎来了癌症集群演化研究的爆发期。最显著的代表就是国际癌症研究组织(ICGC)的引领者Mike Stratton和相关团队针对各个癌症类型的一系列CNS研究文章。我(寸玉鹏